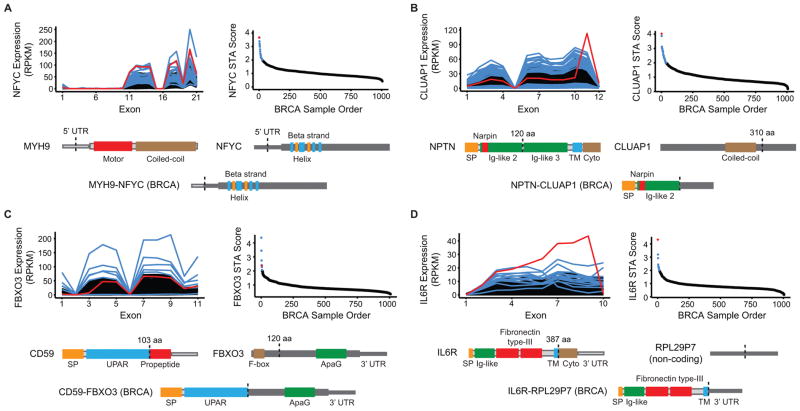
Figure 4. Triple-negative breast cancers harbor a functionally diverse array of gene rearrangements.
A–D, Four examples of STA-predicted rearrangements in triple-negative breast cancers from TCGA. Each panel features an exon-level expression diagram and STA score plot for the gene and cancer type analyzed. Red indicates the representative DNA-validated rearrangement that is depicted at the bottom of each panel as a schematic of the resulting aberrant protein. Blue indicates additional aberrant transcripts meeting STA score threshold. Black indicates background population below threshold. Protein features and untranslated regions (UTRs) are labeled and dotted lines indicate hybrid transcript junctions. aa: amino acid; BRCA: breast invasive carcinoma; Cyto: cytoplasmic domain; nt: nucleotide; SP: signal peptide; TM: transmembrane.