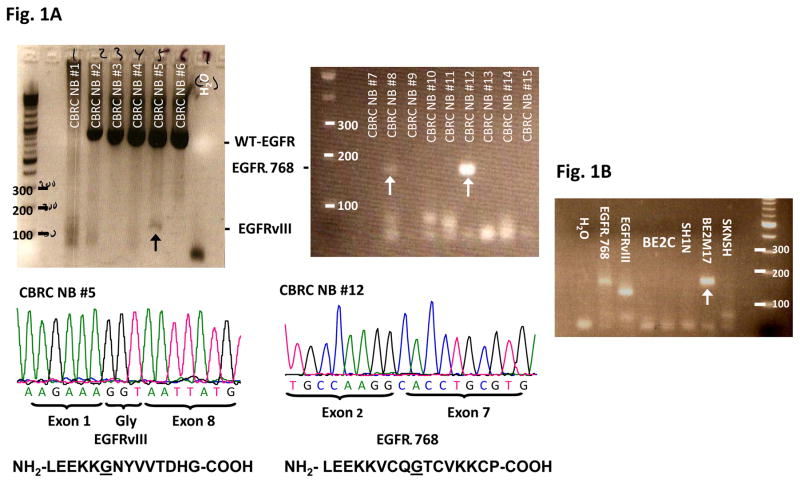
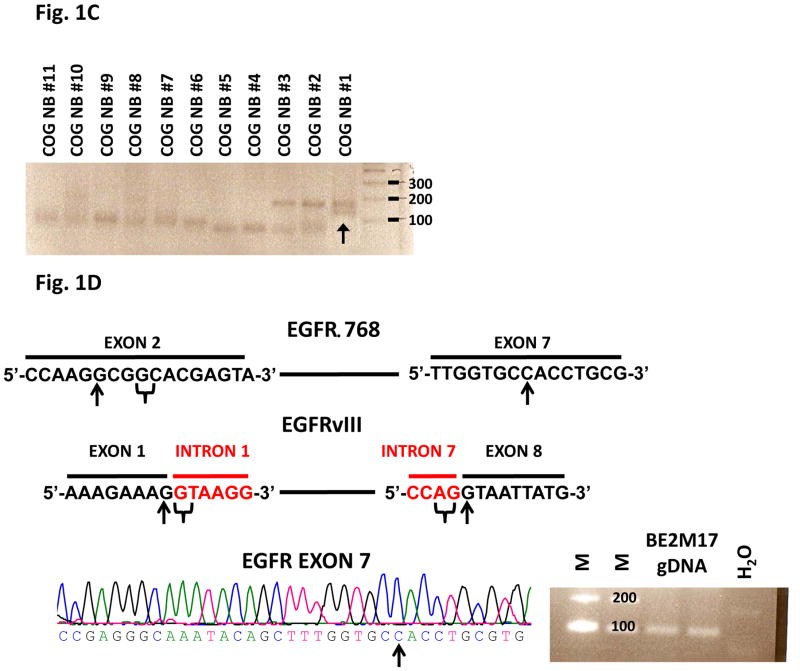
Fig. 1. EGFR extracellular domain mutants in NB.
(A) RT-PCR analysis of EGFR extracellular domain mutations in a representative panel of CBRC NB tumors. Noted that CBRC NB #5 expressed EGFRvIII (black arrow) while CBRC NB #8 and #12 expressed EGFRΔ768 (white arrows). Identities of the bands were confirmed by bidirectional sequencing as shown. The amino acid sequences corresponding to EGFRΔ768 and EGFRvIII were depicted below the nucleotide sequences. The underlined amino acids represented the splice junctions. WT: wild type; Gly: glycine. (B) RT-PCR analysis of EGFR extracellular domain mutations in a panel of NB cell lines. BE2M17 expressed EGFRΔ768 (white arrow). Identity of the band was confirmed by bidirectional sequencing. Gel purified EGFRvIII and EGFRΔ768 cDNA were positive controls. H2O was negative control. (C) RT-PCR analysis of EGFR extracellular domain mutations in a representative panel of COG NB tumors. Noted that COG NB #1 co-expressed EGFRvIII and EGFRΔ768 (black arrow). Identities of the bands were confirmed by bidirectional sequencing. (D) Splice junction analysis of EGFRΔ768 and EGFRvIII. Black arrows pointed to the breakpoints in exon 2/7 of EGFRΔ768 and exon 1/8 of EGFRvIII. The potential/actual splice donor and acceptor sites in the EGFR sequences were bracketed. EGFR exon 7 of BE2M17 genomic DNA was amplified and sequenced as shown in the bottom panel. M: marker; H2O was negative control.