Abstract
Purpose
The initiation, progression, and maintenance of pancreatic ductal adenocarcinoma (PDAC) results from the interplay of genetic and epigenetic events. While the genetic alterations of PDAC have been well characterized, epigenetic pathways regulating PDAC remain, for the most part, elusive. The goal of this study was to identify novel epigenetic regulators contributing to the biology of PDAC.
Experimental design
In vivo pooled shRNA screens targeting 118 epigenetic proteins were performed in two orthotopic PDAC xenograft models. Candidate genes were characterized in 19 human PDAC cell lines, heterotopic xenograft tumor models, and a genetically engineered mouse (GEM) model of PDAC. Gene expression, immunohistochemistry, and immunoprecipitation experiments were performed to analyze the pathways by which candidate genes contribute to PDAC.
Results
In vivo shRNA screens identified BRD2 and BRD3, members of the BET family of chromatin adaptors, as key regulators of PDAC tumor growth. Pharmacological inhibition of BET bromodomains enhanced survival in a PDAC GEM model and inhibited growth of human-derived xenograft tumors. BET proteins contribute to PDAC cell growth through direct interaction with members of the GLI family of transcription factors and modulating their activity. Within cancer cells, BET bromodomain inhibition results in down-regulation of SHH, a key mediator of the tumor microenvironment and canonical activator of GLI. Consistent with this, inhibition of BET bromodomains decreases cancer associated fibroblast content of tumors in both GEM and xenograft tumor models.
Conclusions
Therapeutic inhibition of BET proteins offers a novel mechanism to target both the neoplastic and stromal components of PDAC.
Translational Relevance
Pancreatic ductal adenocarcinoma is extraordinarily chemoresistant and the abundant stromal content of these tumors contributes to the ineffective treatment of this disease. Current approaches in the treatment of PDAC are largely ineffective and utilize drugs that target either the neoplastic cells or the stroma of this disease. This study reveals the broad dependence of pancreatic cancer cell lines and tumor models on the activity of the BET family of chromatin adaptors. BET proteins contribute to PDAC biology by regulating multiple key nodal pathways of this disease, including the direct and indirect regulation of GLI, a family of transcription factors that plays key roles in both epithelial and stromal cells of PDAC tumors. Therapeutic inhibition of BET proteins provides a unique opportunity to simultaneously target both the stromal and neoplastic cells of PDAC.
Keywords: Pancreatic ductal adenocarcinoma, BET bromodomain, SHH, GLI, MYC
INTRODUCTION
The overall five-year survival for PDAC is only 6%, nearly two-thirds lower than any other cancer (1). In addition to their extraordinary chemoresistance, PDAC tumors contain a highly desmoplastic stroma that is a major impediment to the treatment of this disease. This stroma not only promotes the aggressive local growth of the tumor and the intrinsic chemoresistance of the cancer cells, but also acts as a physical barrier to effective chemotherapeutic targeting (2). Clearly, future advances in the treatment of PDAC will require therapeutics that are more effective at killing the cancer cells as well as those that effectively target tumor stroma.
The genetic alterations contributing to PDAC pathogenesis have been extensively studied. The predominant mechanism for the formation of PDAC is through the oncogenic conversion of acinar cells (3). Activating mutations in KRAS initiate acinar-to-ductal metaplasia, with mutations in CDKN2A, TP53, and SMAD4 occurring during progression from pre-neoplastic PanIN lesions to invasive cancer (4). While recent large scale sequencing studies have provided a detailed view of the spectrum of mutations present in subsets of PDAC, these efforts have yet to identify genes that may be therapeutically targeted (5,6). Histone modifications and subsequent chromatin remodeling function as master regulators of gene expression. This regulation is largely mediated by the reversible methylation or acetylation of the tails of histone proteins, and enzymes involved in addition and removal of these modifications are associated with a wide variety of cancers (7). There is also a growing body of evidence linking the proteins recognizing these epigenetic marks to the maintenance and progression of cancer. In this regard, mutations in chromatin regulators (e.g. SWI/SNF components) are present in approximately 20% of PDAC tumors; however, these mutations all involve inactivation of tumor suppressors, and do not readily suggest therapeutic approaches.
In this study we employed an in vivo RNAi screen of a library of epigenetic regulators to functionally identify key mediators of PDAC pathogenesis. shRNAs specific for BRD2 and BRD3, members of the BET family of chromatin adaptors, were identified in a dropout screen performed in an orthotopic model of PDAC. BET proteins contain tandem bromodomains that allow for their binding to acetylated lysines on target proteins to regulate gene expression. Pharmacological inhibition of BET bromodomain binding revealed a critical role of this family in the growth of PDAC cell lines in vitro as well as the in vivo growth of tumors. We found that BET family members regulate PDAC tumor cell growth by modulating the expression of MYC and the activity of the GLI family of transcription factors through direct physical interaction. BET proteins also regulate the tumor microenvironment, at least in part, through the regulation of SHH expression and secretion in the cancer cells.
MATERIALS AND METHODS
Cell Culture
PDAC cell lines were cultured in a 1:1 mix of DMEM and Ham’s F-12 media (Mediatech) supplemented with 10% fetal bovine serum (Life Technologies), penicillin (100 U/ml), and streptomycin (100 μg/ml) at 37°C in 5% CO2. Numerically named PDAC cell lines were all established between 2008 and 20012 in our laboratory from xenograft tumors derived from surgically resected PDAC. These cell lines are described in more detail in the supplemental experimental procedures. BxPc3 (2010), COLO357 (2011), CFPAC-1 (2002), and PANC-1 (2002) were obtained from ATCC. Mouse PDAC cell line NB494 (2015) was a generous gift from Nabeel Bardeesy (Massachusetts General Hospital) (8). All cell lines were routinely evaluated for morphology and tested for mycoplasma. No genetic authentication was performed. Experiments evaluating the effects of BET inhibition employed CPI203 (BETi), and its inactive enantiomer CPI440 or DMSO as controls.
Tumor models
Orthotopic tumors were formed by injecting PDAC cell lines 1312 and 1275 (1×106 cells) into the pancreata of 6 week old nu/nu mice (Jackson Laboratory). Heterotopic xenograft tumors were established by subcutaneous injection of PDAC cells (1–3×106 cells) in nu/nu mice. The Pdx1-Cre (9), LSL-KRASG12D (10), and p53flox/flox (11) were bred to generate the Pdx1-Cre;LSL-KRASG12D;p53−/− mice. Starting at six weeks of age, mice were treated with BETi or vehicle control and remained in the study until reaching a moribund condition or spontaneous death.
Gene expression analysis
PDAC cells were exposed 1.6 μM BETi or DMSO for 5 or 10 hours. Total RNA was isolated from cells and analyzed using HumanHT-12 v4 Expression BeadChips (Illumina) by the Laboratory of Molecular Medicine at the Partners Healthcare Center for Personalized Genetic Medicine. This gene expression data has been deposited in the NCBI Gene Expression Omnibus (accession number GSE55209).
Additional experimental details are presented in the Supplemental Experimental Procedures
RESULTS
Identification of BET proteins in pancreatic tumorigenesis
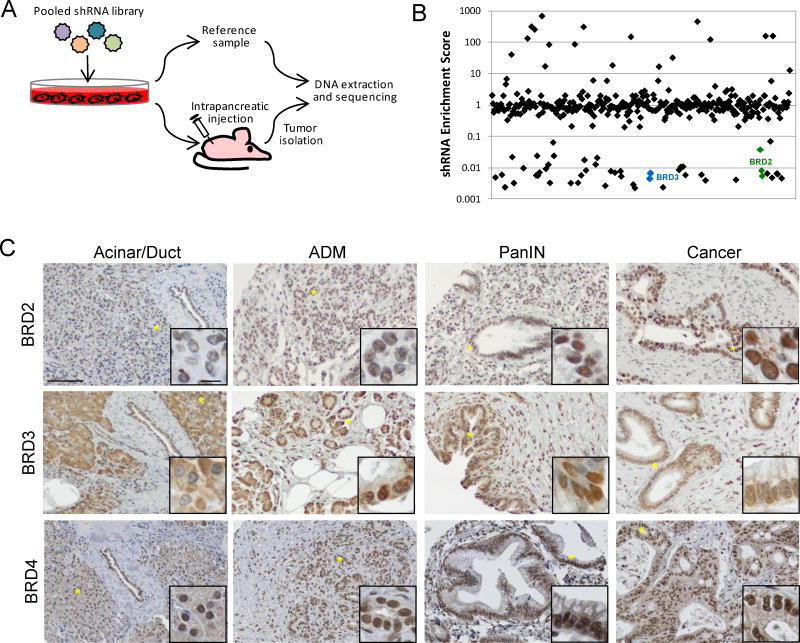
We sought to functionally identify epigenetic regulators contributing to PDAC by conducting pooled shRNA screens. Since epigenetic programs are altered when PDAC cells are cultured in vitro, we performed these screens in vivo using low passage patient-derived cell lines implanted as orthotopic tumor models (12). These studies further allowed for the identification of those factors important for tumor growth in a microenvironment that closely recapitulates the human disease. The screens employed 406 shRNAs that targeted 118 genes from three general classes of proteins: histone methyltransferases, histone demethylases, and chromatin adaptors (Supplementary Fig. S1A). A pooled lentiviral library expressing these shRNAs was used to infect PDAC cell lines and 24–48 hrs after infection, cells were implanted into pancreata of mice and allowed to form orthotopic tumors (Fig. 1A). Depleted shRNAs were identified by comparing the relative abundance of an shRNA in the tumor to that of the original population of infected cells (Fig. 1B, Supplementary Fig. S1B and C, and Supplementary Table S1). Identified in this screen were BRD2 and BRD3, members of the BET family of chromatin adaptor proteins. At least two shRNAs targeting each of these genes were depleted more than 15-fold in tumors derived from each cell line.
Figure 1.
BET proteins are identified in an in vivo shRNA screen for epigenetic regulators in PDAC. A. Schematic representation of the pooled shRNA screen. B. Representative shRNA distribution from a tumor derived from cells infected with pooled shRNA library. Individual shRNAs are plotted on the x-axis and their relative enrichment/depletion in the tumor is plotted in log scale on the y-axis. Depleted shRNAs targeting BRD2 (green) and BRD3 (blue) are indicated. C. Representative images of normal, premalignant, and malignant pancreatic tissue stained with antibodies specific for BRD2, BRD3, and BRD4. Scale bars = 100 μm in main images and 10 μm in insets. Locations of the inset images within the main images are indicated by asterisks.
To begin to understand the role of BET family members in PDAC tumorigenesis, their expression in normal pancreas and malignant tissue was analyzed employing a tissue microarray containing 121 surgically resected pancreatic specimens, including 91 unique PDAC samples (Fig. 1C and Supplementary Fig. S2) (13). Consistent with their identification in the in vivo shRNA screen, elevated nuclear expression of BRD2 and BRD3 was observed in pre-malignant and malignant tissue relative to normal pancreas. Weak nuclear expression of BRD2 was observed in the vast majority of normal acinar cell samples. Upon undergoing acinar-to-ductal metaplasia (ADM), cells expressed higher nuclear BRD2 levels (p<0.013) that were comparable to that observed in normal ductal epithelium. This increase in BRD2 levels was maintained in primary and metastatic tumors (n=121, p<0.003). Nuclear expression of BRD3 was rarely observed in normal acinar cells but was strongly expressed in the nuclei of cells undergoing ADM (p<0.001) and cells throughout PanIN progression (n=22, p<0.001) to primary and metastatic disease (n=92, p<0.004). In contrast to BRD2 and BRD3, nuclear BRD4 expression was readily detected in normal acinar and duct cells, as well as cells throughout the histological progression to cancer. No significant differences in the staining of BRD4 were observed between these samples.
Pharmacological inhibition of BET proteins impairs PDAC cell growth
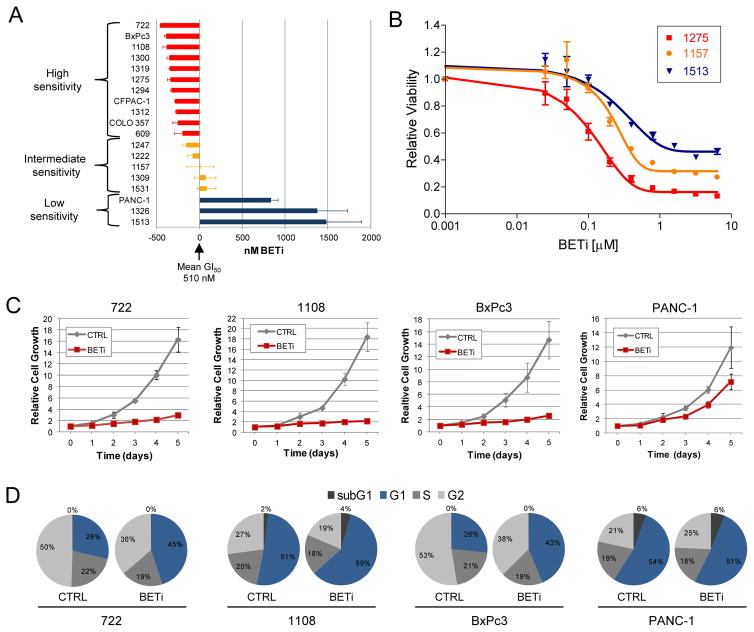
To evaluate the contribution of BET proteins to in vitro growth of PDAC cells, we employed the BET bromodomain inhibitor CPI203 (BETi). This small molecule acts by competitively inhibiting the binding of BET bromodomains to acetylated lysines (14). Nineteen PDAC cell lines, including 15 low-passage cell lines derived from xenograft tumors, were treated with increasing amounts (0–6.4 μM) of BETi to evaluate their effects on cell growth (Fig. 2A). Eighty-four percent of the cell lines tested were responsive to BET bromodomain inhibition (GI50 < 1 μM) (Supplementary Fig. S3A), but not to treatment with the inactive enantiomer, CPI440 (data not shown). PDAC cells had a mean GI50 of 510 nM and, based on their deviation from the mean, cell lines were defined as exhibiting high (GI50 < 350 nM), intermediate (GI50 < 600 nM), or low sensitivity to BETi (GI50 > 1 μM). Similar differences in the effects of BETi on high and low sensitivity cells were observed with a second BET bromodomain inhibitor, JQ1 (Supplementary Fig. S3B). These groups generally exhibited an inverse correlation between BETi sensitivity and the percentage of residual cell viability observed at the maximum dose of BETi (Fig. 2B), likely reflecting differing heterogeneity of the cell populations. Sensitivity to BET bromodomain inhibition between the cell lines appeared to be independent of relative BRD2, BRD3, or BRD4 expression levels (Supplementary Fig. S3C).
Figure 2.
BET bromodomains contribute to PDAC cell growth in vitro and in vivo. A. PDAC cell lines were exposed to increasing amounts (0–6.4 μM) of BETi and analyzed for changes in growth after six days. The deviation from the mean GI50 of each cell line is shown. B. Representative dose response curves for cell lines exhibiting high (red), intermediate (orange), and low (blue) sensitivity to BETi are shown. C. Growth curves of PDAC cell lines treated with 1.6 μM BETi or CPI440 (CTRL). The averages (±SEM) from at least two experiments performed in duplicate are shown. D. Distribution of PDAC cells in the cell cycle was determined by propidium iodide staining after treatment with 1.6 μM BETi or CPI440 (CTRL) for three days. Results are representative of three experiments.
To further interrogate the effects of BET bromodomain inhibition, the growth kinetics of high and low sensitivity cell lines were analyzed (Fig. 2C). High sensitivity cell lines (722, 1108, and BxPc3) exhibited a nearly five-fold growth difference after five days whereas low sensitivity PANC-1 showed only a 2-fold reduction in growth. Increases in apoptosis were not observed in cells after treatment with BETi (data not shown). However, cell cycle analysis revealed BETi induced an increase in the proportion of cells in G1 in high sensitivity cell lines, whereas the cell cycle distribution in low sensitivity cells was not altered (Fig. 2D). Analysis of regulators of the G1 to S transition revealed increased abundance of p27 Kip1, a negative regulator of cyclin dependent kinases (CDKs), in high sensitivity cells treated with BETi (Supplemental Fig. S3D). Consistent with this, levels of phosphorylated Rb (Ser780) were decreased in these cells. Interestingly, total levels of Rb were also diminished by BETi, suggesting a secondary mechanism by which BET bromodomain inhibition affects cell cycle progression. In accordance with our cell cycle analysis, these alterations in p27 Kip1, phospho-RB, and total Rb were not observed in low sensitivity PANC-1 cells.
BET regulation of MYC contributes to the biology of PDAC
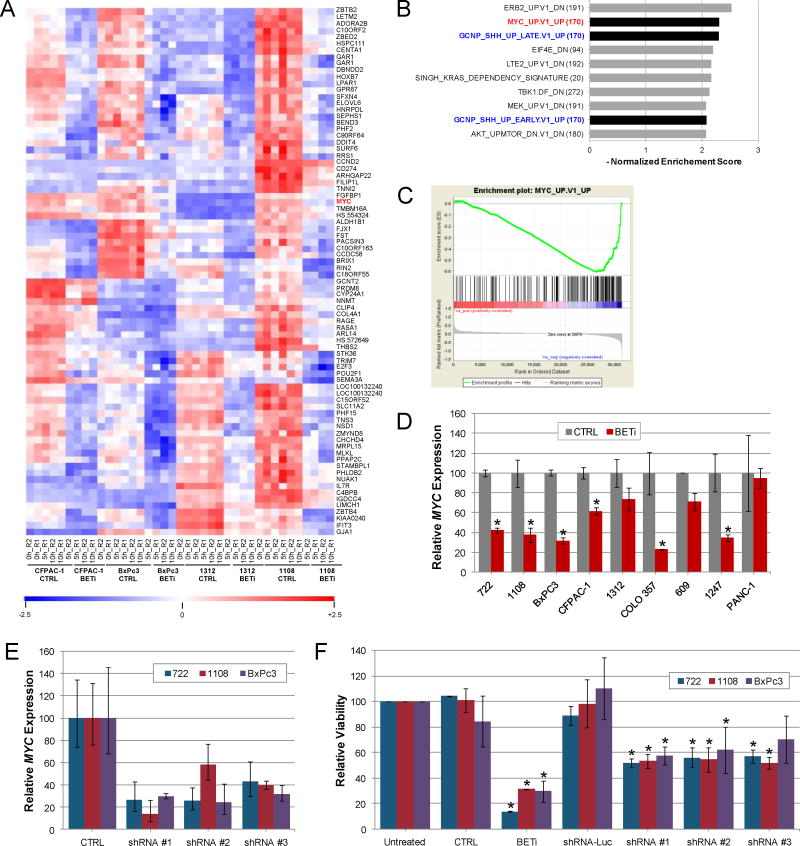
To determine the mechanism(s) by which BET proteins contribute to PDAC cell growth, gene expression profiling was performed after 5 and 10 hours of BET bromodomain inhibition in four cell lines responsive to BETi (CFPAC-1, BxPc3, 1312, and 1108). Among the top 80 genes showing significant down-regulation (≥1.75 fold, p<0.05) was MYC, a known target of BET proteins (Fig. 3A). In addition to decreased expression of MYC, gene set enrichment analysis (GSEA) revealed that one of the top gene sets down-regulated by BET inhibition involved the MYC pathway (Fig. 3B and C and Supplemental Fig. 4A). qPCR analysis of cells exposed to BETi demonstrated a significant reduction in MYC in 6 of 8 cell lines with high or intermediate sensitivity to BETi, and lack of effect in the low sensitivity PANC-1 (Fig 3D). These results are consistent with previous observations that BET proteins regulate MYC expression in a subset of PDAC cells (15,16).
Figure 3.
Regulation of MYC by BET family members contributes to growth of PDAC cells. A. Genome wide transcription analysis of PDAC cell lines. A heat map of the top 80 down-regulated genes in CFPAC-1, BxPc3, 1312, and 1108 cells after BET bromodomain inhibition is shown. High (red) and low (blue) gene expression are indicated. B. Top 10 programs identified by GSEA analysis (C6) of BET-regulated genes. C. GSEA enrichment plots comparing BETi responsive genes to those regulated by MYC. D. qPCR analysis of MYC expression in PDAC cell lines treated with BETi for 10 hours. Average (± std. dev.) is shown. Asterisk indicates changes in gene expression with p<0.05. E. Expression of MYC in PDAC cells after infection of viruses expressing control shRNA (CTRL) targeting luciferase or shRNAs targeting MYC. qPCR analysis of MYC was normalized to uninfected cells and the average and standard deviation is shown. F. PDAC cell lines infected with shRNAs were analyzed for changes in cell viability six days after infection. Parallel cultures of uninfected cells were treated with 1.6 uM BETi or control. Cell viability was normalized to uninfected cells and the average and standard deviation is shown. Asterisk indicates changes in viability with p<0.05.
To support the potential role of MYC inhibition in the response to BETi, we tested the impact of three shRNAs targeting MYC on PDAC cell growth (Fig. 3E). Each MYC-specific shRNA reduced the proliferation of three high sensitivity cell lines, although the effects were considerably weaker than BETi (Fig. 3F). Thus, these results demonstrate a significant role for MYC in the biology of PDAC cells and implicate MYC suppression in the response to BET bromodomain inhibition while also suggesting the existence of other important downstream targets.
BET proteins regulate GLI activity in PDAC
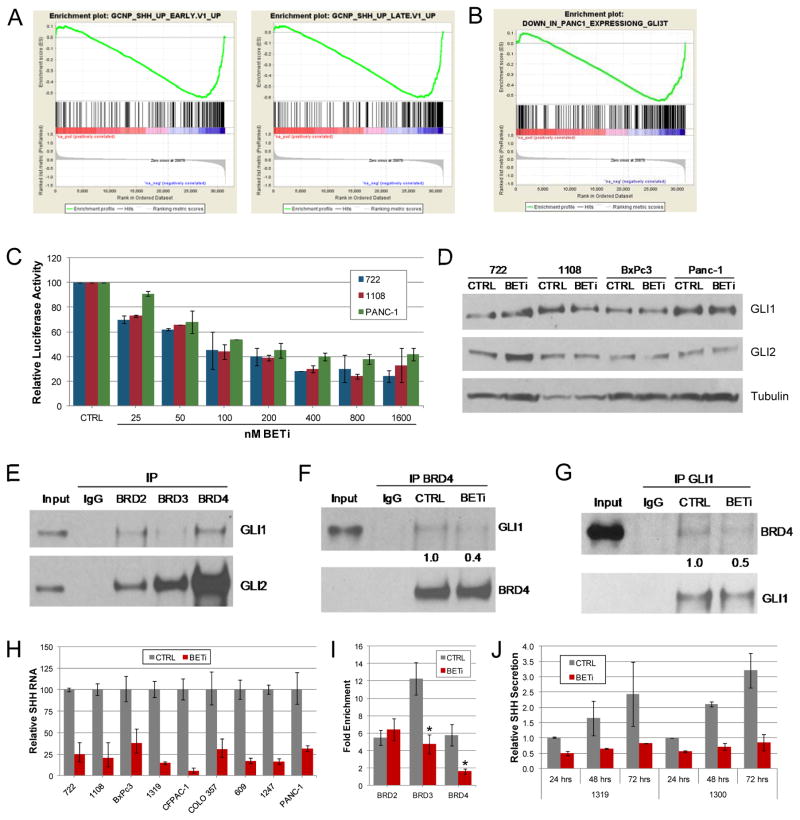
Our GSEA also suggested a prominent role for BET proteins in the regulation of the SHH-GLI signaling pathway (Fig. 3B and Fig. 4A). This pathway has dual roles in PDAC, cell autonomous functions for GLI proteins in PDAC cell growth, and a distinct paracrine function of SHH produced by PDAC cells in the proliferation of the stroma (17,18). Therefore, the SHH-specific gene sets identified in our GSEA likely reflect SHH-independent alterations in GLI activity in PDAC cells. Consistent with this hypothesis, a significant overlap between BET-responsive genes and those dependent on GLI activity was observed (Fig. 4B and Supplemental Fig. S4B) (18). The BET-dependent expression of a subset of these GLI target genes was validated by qPCR in PDAC cell lines (Supplementary Fig. S4C). Importantly, these reductions in gene expression correlated with decreased GLI activity in PDAC cells. Employing a luciferase reporter vector driven by a GLI-dependent promoter, we observed greater than 60% decrease in GLI-luciferase activity after 24 hours of BET bromodomain inhibition in four PDAC cell lines (Fig. 4C and Supplementary Fig. S4D and E). Additionally, this change in GLI activity exhibited a dose-dependent response to BETi, with as little as 100 nM of BETi suppressing GLI-luciferase activity by 50% or more. These results demonstrate that BET proteins play a key role in maintaining the activity of GLI in PDAC cells and diminished GLI activity likely contributes to the biological effects of BETi.
Figure 4.
BET bromodomain proteins regulate the activity of GLI in PDAC cells. A. GSEA enrichment plots comparing BETi responsive genes to those regulated by SHH. B. GSEA enrichment plot comparing BETi responsive genes to those regulated by GLI in PDAC cells. C. Relative luciferase activity of a GLI-dependent luciferase reporter construct containing 9 tandem GLI binding sites in PDAC cell lines after 24 hours of treatment with increasing concentrations (25–1600 nM) of BETi or 1600 nM control (CPI440). The averages (± SD) from 2 independent experiments are shown. D. Western blot of GLI1 and GLI2 in PDAC cells treated with 1.6 μM BETi or control (CPI440) for 24 hours. E. Immunoprecipitation experiments performed with control IgG or antibodies specific to BRD2, BRD3, and BRD4 and cell lysates from PANC-1 cells ectopically expressing GLI1. Representative Western blots detecting the co-immunoprecipitation of GLI1 (top) and GLI2 (bottom) are shown. F. Immunoprecipitations with cell lysates from PANC-1 cells ectopically expressing GLI1 were performed with control IgG and antibodies specific for BRD4 in the presence of BETi or control CPI440. Representative Western blots detecting co-precipitated GLI1 (top) and precipitated BRD4 (bottom) are shown. Immunoprecpitated BRD4 and co-precipitated GLI1 were quantified and their ratio normalized to that observed in immunoprecipitations containing CPI440. G. Reciprocal immunoprecipitation-western blot experiments to those described in panel F detecting the co-precipitation of BRD4 (top) and precipitation of GLI1 (bottom). H. qPCR analysis of SHH expression in PDAC cell lines treated with BETi or vehicle control for 10 hours. I. The presence of BRD2, BRD3, and BRD4 on the endogenous SHH promoter in PANC-1 cells was determined by chromatin immunoprecipitation. The average (± SEM) fold enrichment relative to control IgG of two independent experiments are shown. Asterisks indicate significant (p<0.03) changes in binding of after BET inhibition. J. Secretion of SHH from PDAC cell lines was determined by ELISA after exposure to BETi or control (CPI440) for the indicated times.
To understand the mechanisms by which BET proteins regulate the activity of GLI, we examined the effects of BETi on the expression GLI. qPCR analysis revealed dramatic decreases in GLI1 mRNA levels in the majority of cell lines exposed to BETi for 24 hours (Supplementary Fig. S5A). GLI2 mRNA levels were also diminished in half of these cell lines. Greater than 50% reductions in GLI1 and GLI2 mRNA are seen between 4–8 hours of BET bromodomain inhibition (Supplementary Fig. S5B). Despite this, little to no decrease in their protein levels occurred after 24 hours (Fig. 4D), a time by which GLI-luciferase activity and expression of GLI target genes are diminished. Protein half-life studies revealed that the discrepancy between GLI RNA and proteins levels was due to an increased GLI protein half-life after BET bromodomain inhibition. While GLI1 and GLI2 half-life is 4–8 hours in control cells, little change in their protein levels are observed in BETi treated cells even after 24 hours of protein synthesis inhibition (Supplementary Fig. S5C). Taken together, these results suggest that suppression of the GLI transcriptional program in PDAC cells by BET bromodomain inhibition is not due to reduced expression of GLI1 and GLI2.
The lack of significant changes in the proteins levels of GLI1, GLI2, and GLI3 (Supplementary Fig. S5D) after BET bromodomain inhibition suggested that the observed reductions in GLI target gene expression and GLI-luciferase activity is due to a direct regulation of GLI by BET proteins. Co-immunoprecipitation experiments were performed to determine whether BET and GLI proteins are found in the same protein complexes. In PDAC cells ectopically expressing GLI1, immunoprecipitations of BRD2, BRD3, and BRD4 resulted in the co-precipitation of GLI1 (Fig. 4E). Reciprocal experiments using antibodies specific to GLI1 identified the co-precipitation BRD4 (Supplementary Fig. S6A). Furthermore, this interaction between GLI1 and BRD4 was diminished by BET bromodomain inhibition (Fig. 4F and G), indicating the transcriptional activation activity of GLI1 is regulated by the physical association of BET proteins. Similar to GLI1, co-immunoprecipitation experiments demonstrated the presence of GLI2 in complexes containing BRD2, BRD3, and BRD4 (Fig. 4E and Supplementary Fig. S6B and C). Although the interaction of GLI2 and BRD4 could be detected with endogenous expression of these proteins (Supplementary Fig. S6D), these complexes were refractory to BET bromodomain inhibition (Supplementary Fig. S6E), suggesting that GLI1 and GLI2 interact with BET proteins through distinct mechanisms. Taken together, these results support GLI as key component of the BET-dependent growth program in PDAC
Given the important role of SHH in activating the GLI transcriptional program in the tumor stroma, we investigated whether BETi so alters SHH expression. qPCR analysis revealed the expression of SHH was reduced by 60% or more in PDAC cells after BET bromodomain inhibition (Fig. 4H). Consistent with the paracrine function of SHH in PDAC, the extent by which SHH expression was reduced in cells was independent of the biologic sensitivity of cells to BETi. Chromatin immunoprecipitation experiments revealed that BET proteins are bound to the SHH promoter in PDAC cells and binding of BRD3 and BRD4 is diminished after BET bromodomain inhibition (Fig. 4I), suggesting a direct role for BET proteins in the regulation of SHH expression. Furthermore, the decreased expression of SHH RNA corresponded to a sustained decrease in SHH secretion as determined by ELISA (Fig. 4J). Taken together, these results suggest that BET proteins regulate multiple components of the SHH-GLI pathway in PDAC cells and BET bromodomain inhibition may influence PDAC tumors by affecting neoplastic cells through inhibition of GLI and the tumor stroma through decreased expression of SHH.
BET bromodomain activity is required for PDAC tumor growth
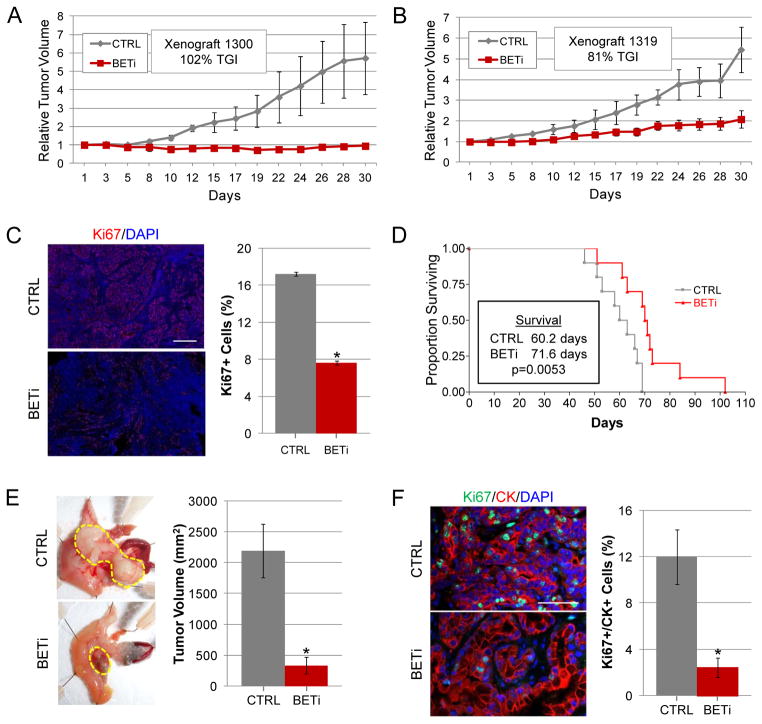
To directly examine whether BET proteins are required for the growth of PDAC tumors in vivo, we evaluated the effects of BETi on the growth of xenograft tumors derived from human PDAC cell lines. Mice bearing heterotopic tumors were treated with BETi (10 mg/kg) or vehicle control twice daily using a 5-days on;2-days off dosing regimen. On average, tumors in control mice continued to grow to five times their starting size while tumors in mice treated with BETi exhibited little to no growth (Fig. 5A, p<0.05 and 5B, p<0.03). Importantly, immunostaining with a human ki67-specific antibody revealed a 60% reduction (p<0.001) in proliferation of the human cancer cells in BETi treated xenograft tumors relative to controls (Fig 5C), while no differences in histology or apoptosis were observed (Supplemental Fig. S7A and data not shown). Thus, PDAC cells are highly responsive to BETi in vivo.
Figure 5.
BET bromodomains regulate PDAC tumor growth. A–B. Mice bearing heterotopic tumors derived from the PDAC cell line 1300 or 1319 were treated for 30 days with BETi (1300 n=3; 1319 n=5) or vehicle control (1300 n=3; 1319 n=3) upon tumors reaching 125 mm3. Average relative tumor volumes (±SEM) are plotted. Percent tumor growth inhibition (TGI) relative to control mice is indicated. C. Tumors derived from the PDAC cell line 1300 treated with BETi or vehicle control were analyzed for their proliferative index by staining for Ki67. Representative images from tumors are shown. Scale bars = 100 μm. Quantitative analysis of Ki67 stained sections are shown on the right. Asterisk indicates p<0.0001. D. A Kaplan-Meier survival plot of genetically engineered mice (Pdx-1-Cre;KRASG12D;p53−/−) treated with BETi (n=10) or vehicle control (n=10) starting at 6 weeks of age. E. Representative pictures of tumor bearing pancreata from control and BETi treated mice. The borders of the tumors are outlined in yellow. The average (±SD) tumor volumes are plotted on the right. Asterisk indicates p<0.003. F. Tumors derived from the Pdx-1-Cre;KRASG12D;p53−/− mice treated with BETi or vehicle control were analyzed the expression of Ki67 and cytokeratin by immunofluorescence. Representative images from tumors are shown. Scale bars = 100 μM. Quantitative analysis of cytokeratin and Ki67 stained sections are shown on the right. Asterisk indicates p<0.02.
To further examine the potential of BETi as a PDAC therapy, we employed a GEM model (Pdx-1-Cre;KRASG12D;p53−/−). A cell line derived from one of these tumors exhibited decreased expression of MYC (53%, p<0.001) and SHH (85%, p<0.001), as well as diminished GLI transcriptional activity (39%, p<0.002) in response to BETi, indicating that BET proteins regulate these core pathways in both human and mouse PDAC (Supplemental Fig. S8). Pdx-1-Cre;KRASG12D;p53−/− mice form highly aggressive PDAC by 6 weeks of age and have an average survival of 60 days (Fig. 5D) (8). Administering BETi to these mice after tumor formation (at 6 weeks of age) prolonged their average survival to more than 71 days (p<0.006). Notably, at the time of necropsy, tumors from BETi treated mice were six times smaller (p<0.003) than those from control mice (Fig. 5E). Despite this dramatic reduction in tumor burden, the death of BETi-treated mice appeared to be cancer related, as tumors from BETi treated mice frequently obstructed the intrapancreatic bile duct and non tumor-bearing mice treated with BETi for 28 days did not exhibit evidence of cytotoxicity (data not shown). Consistent with that observed for xenograft tumors, an 80% decrease (p<0.02) in the proliferation of cytokeratin-positive tumor cells was observed in BETi treated mice relative to controls (Fig. 5F). There was no appreciable difference in the histology of tumors from control and BETi treated mice as tumors in both groups were moderate to poorly differentiated and contained cells with a mesenchymal morphology (Supplementary Fig. S7A).
BET bromodomain activity regulates key pathways for PDAC tumor growth and maintenance of the tumor microenvironment
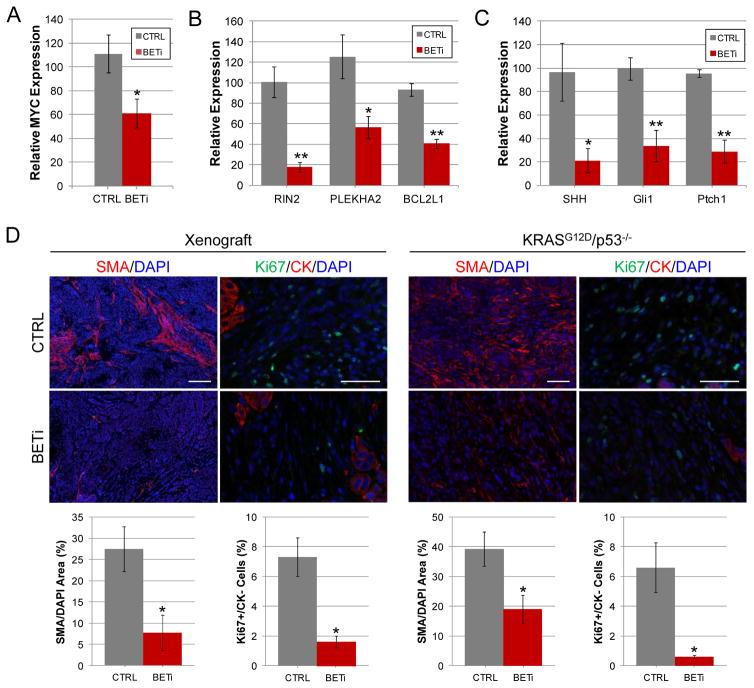
Given the robust effect of BET bromodomain inhibition on the growth of PDAC tumor models, we examined whether the BET-regulated pathways identified in PDAC cell lines are also altered in tumors. Mice bearing orthotopic PDAC tumors were treated with BETi or vehicle control for three days, and RNA from tumors were examined for changes in gene expression. The expression of MYC was reduced by 45% (p<0.05) in mice treated with BETi relative to control mice. Additionally, the expression of GLI target genes was decreased 54–80% (p<0.03) after BET bromodomain inhibition, consistent with a reduction in GLI activity in the cancer cells. Furthermore, an 80% decrease (p<0.04) in SHH was observed in the human-derived cancer cells from mice treated with BETi relative to those from control mice (Fig. 6C). Consistent with suppression of SHH signaling, the expression of canonical hedgehog target genes, Gli1 and Ptch1, were decreased by 70% (p<0.008) in the mouse-derived tumor stroma. These results indicate that BET proteins contribute to the regulation of the MYC, GLI, and SHH programs of PDAC in vitro and in vivo.
Figure 6.
BET bromodomains regulate key pathways required for PDAC cell growth and maintenance of the tumor microenvironment. A. qPCR analysis of MYC expression in neoplastic cells of orthotopic tumors formed by the implantation of human PDAC cell line 1319 is shown. Asterisk indicates significant (p<0.05) change in gene expression. B. qPCR analysis of RIN2, PLEKHA2, and BCL2L1 expression in neoplastic cells of 1319 orthotopic tumors is shown. Single and double asterisks indicate significant (p<0.03 and p<0.001, respectively) changes in gene expression. C. qPCR analysis of SHH expression in neoplastic cells of 1319 orthotopic tumors and Gli1 and Ptch1 expression in the mouse-derived stroma are shown. Single and double asterisks indicate significant (p=0.03 and p<0.008, respectively) changes in gene expression. D. Heterotopic tumors derived from the PDAC cell line 1300 and KRASG12D;p53−/− mice treated with BETi or vehicle control as described in Fig. 5 were analyzed for their tumor associated fibroblast (SMA) and proliferation Ki67 of cytokeratin negative stromal cells. Representative images from tumors are shown. Scale bars for SMA and Ki67 images = 100 μm and 50 μm respectively. Quantitative analysis of SMA and Ki67stained sections are shown below the corresponding images. Asterisk indicates p<0.04.
Since the SHH produced by neoplastic cells promotes proliferation of cancer associated fibroblasts via paracrine signaling, we hypothesized that BET proteins might regulate the fibroblast content of the tumor stroma by altering the expression of SHH. Therefore we examined the effects of long-term BET bromodomain inhibition on the tumor microenvironment using both human cell line-derived xenograft and GEM models of PDAC. Three major components of the tumor stroma, collagen, cancer associated fibroblasts, and tumor vasculature were analyzed using Mason-trichrome staining and antibodies specific for smooth muscle actin (SMA) and CD31. No significant differences in collagen content or CD31-positive blood vessels were observed between control and BETi treated tumors (Supplemental Fig. S7B). However, in agreement with the decreased SHH signaling observed above, the fibroblast content of tumors treated with BETi was more than 2-fold lower (p<0.03) in Pdx-1-Cre;KRASG12D;p53−/− mice and 4-fold lower (p<0.05) in xenograft models relative to control mice (Fig. 6D). Ki67 staining of cytokeratin negative cells revealed tumor stromal cell proliferation was more than 3-fold lower (p<0.02) in xenograft models and 10-fold lower (p<0.03) in tumors from Pdx-1-Cre;KRASG12D;p53−/− mice treated with BETi relative to control mice. While these results correlated with decreased expression of SHH in the cancer cells, BET proteins may also play a direct role in the tumor stroma. BRD2, BRD3, and BRD4 are expressed in the stroma of xenograft tumors (Supplemental Fig S7C) which is consistent with the expression of BET proteins in the cancer associated fibroblasts of human PDAC samples (Supplemental Fig S7D). Nevertheless, these results define an important role for BET bromodomain activity in the maintenance of the cancer associated fibroblasts in both murine and human tumor models of PDAC.
BRD2 and BRD4 regulate the SHH-GLI pathway
To evaluate the contribution of individual BET proteins to the effects of BET bromodomain inhibition, we employed shRNAs targeting BRD2, BRD3, and BRD4 in two low-passage cell lines that were highly sensitive to BETi, 722 and 1108. Each shRNA effectively diminished the expression of the corresponding BET protein in these cells (Supplemental Fig S9A and B). Western blots analyzing the effects of two shRNAs targeting each BET family member revealed a key role for BRD2, BRD3, and BRD4 in the expression of MYC (Supplemental Fig S10A and B). However, one BRD2-specific shRNA (5240) only had modest effects on MYC expression in 722 cells. While each BET protein contributes to the expression of MYC, BRD2 and BRD4 are the only BET family members that contribute to the regulation of SHH and GLI. Cells with diminished BRD2 or BRD4 levels exhibited reduced expression of SHH (29–57%, p<0.03; Supplemental Fig. S10C and D). Further, these cells had lower Gli-luciferase activity (29–72%, p<0.04; Supplemental Fig. S10E and F) and reduced levels of the GLI target gene, PLEKHA2 (17–43%, p<0.03; Supplemental Fig. S10G and H). Corresponding changes were not observed in cells with diminished levels of BRD3. Taken together, these results implicate BRD2 and BRD4 in the regulation of MYC, SHH, and GLI, and suggest a unique role for BRD3 in the regulation of MYC.
Analysis of the growth of cells expressing the BET-specific shRNAs suggested the relative importance of individual BET proteins was dependent on the cell line analyzed. The growth of 722 cells was selectively diminished (21–26%, p<0.05) by the expression of shRNAs targeting BRD2 (Supplemental Fig. S11A). In contrast, expression of shRNAs targeting BRD4 reduced the growth of 1108 cells by 32–52% (p<0.001, Supplemental Fig S11B). To further interrogate the effects of reduced BET expression, 1108 cells expressing BET-specific shRNAs were implanted into immunodeficient mice (n=3 for each shRNA). The ability of these cells to form heterotopic tumors was compared to cells expressing a non-targeting control shRNA implanted into the opposite flank of each animal. In contrast to the selective role for BRD4 in the growth of these cells in vitro, reduced expression of each BET protein resulted in at least a 65% reduction (p<0.02) in tumor growth, with no changes in histology (Supplemental Fig S11C–E). Analysis of Ki67 staining of these tumors did not reveal a reduction in cancer cell proliferation, suggesting the reduced tumor size was due to a delayed tumor initiation rather than a slower growth of the cancer cells (Supplemental Fig S12A and B). In accordance with the diminished expression of SHH in these cells in vitro, the abundance of SMA-positive cancer associated fibroblasts were reduced by at least 45% (p<0.02) in tumors formed from cells expressing each shRNA targeting BRD2 (Supplemental Fig S12A and C). A similar trend was observed with tumors derived from cells expressing both shRNAs targeting BRD4, although one shRNA failed to reach significance (shRNA 1277, p<0.009; shRNA 1532, p<0.08). These results suggest BRD2 and BRD4 regulate the tumor microenvironment through paracrine signaling pathways.
DISCUSSION
Employing an in vivo functional RNAi screen of epigenetic regulators, we identified a new epigenetic pathway controlling the biology of PDAC. The BET family of chromatin adaptors contributes to both compartments of PDAC tumors by influencing multiple key nodal pathways, including MYC, GLI, and SHH. BET proteins represent a novel class of transcriptional coactivators of GLI, a family of transcription factors that play key roles in the biology of PDAC cells and their associated tumor stroma.
Here we demonstrated that GLI transcriptional activity is dependent on BET bromodomain activity in PDAC cells. GLI1 and GL2 physically interact with BRD2, BRD3, and BRD4, defining a novel mechanism for the regulation of GLI activity. Since BET proteins have been found to be components of the super elongation complex and polymerase-associated factor complex, it is likely that they contribute to GLI transcriptional activity by mediating the release of paused RNA PolII on the promoters of GLI target genes (19). Acetylated lysines in both GLI1 (K518) and GLI2 (K757) regulate their transcriptional activity (20,21). However, only the interaction of GLI1 and BET proteins was sensitive to BETi, indicating that GLI1-BET complexes are largely responsible for the bromodomain-dependent GLI activity observed in PDAC cells and suggests a bromodomain-independent mechanism by which BET proteins associate with GLI2. Decreases in GLI activity after BET bromodomain inhibition have also been reported in mouse fibroblasts and mouse models of medulloblastoma (22,23). However these changes in GLI activity involve the direct regulation of GLI mRNA expression and ultimate control of corresponding protein levels by BET proteins. In PDAC cells, GLI protein levels remain unchanged after short-term BET bromodomain inhibition due to an extended protein half-life. While the mechanism for this increased protein stability is unknown, it is possible that GLI exhibits decreased stability when it is part of BET complexes to allow for tighter transcriptional regulation of GLI target genes. While this increased protein stability is unlikely to overcome the reduction in GLI RNA with long-term BET bromodomain inhibition, these studies have revealed the physical interactions between these families of proteins and provide insight into their normal function in the regulation of transcriptional pathways important for PDAC.
The significance of the GLI transcriptional program to PDAC has recently become apparent. GLI activity in neoplastic cells of PDAC is independent of canonical SHH signaling (24). The mechanism by which GLI is activated in PDAC cells is unclear, but involves KRAS and TGFβ (17,25). Inhibition of GLI activity by shRNA or a dominant negative mutant of GLI (Gli-3T) in human PDAC cells resulted in reduced soft agar colony formation (17,25). Incorporation of Gli-3T in mouse models of PDAC revealed that GLI activity is required for the formation of premalignant PanIN lesions in KRASG12D mice and the formation of PDAC tumors in KRASG12D;p53+/− mice (18). In addition to its role in cancer cells, GLI is a key factor within the tumor stroma, where it is activated by canonical SHH signaling. SHH is secreted by the neoplastic cells of PDAC tumors and stimulates cancer-associated fibroblasts via paracrine signaling to promote formation of the tumor desmoplasia (24,26). This event begins very early in the development of PDAC. SHH is not generally expressed in the adult pancreas but is activated during the development of pre-neoplastic PanIN lesions (27). Studies employing GEM models of PDAC have demonstrated the progressive appearance of a desmoplastic and pro-inflammatory stroma during the progression from PanIN to PDAC (28,29). A key role for SHH in development of the tumor stroma was recently confirmed in Pdx1-Cre;KRASG12D;p53+/− mice that lack SHH expression; tumors from these mice have a greatly diminished fibroblast and immune infiltrate relative to tumors formed from mice expressing SHH (30). Although we cannot exclude a direct role for BET proteins in regulating the tumor stroma, the reduced fibroblast content of BETi treated tumors corresponded with decreased expression of SHH in PDAC cells after pharmacological or shRNA-mediated reduction in BET protein activity.
The role of the SHH-dependent stroma in PDAC has been somewhat controversial. Initial studies inhibiting SHH signaling demonstrated a collapse of the tumor stroma, enhanced tumor vascularity, and increased delivery of gemcitabine to tumors (31). This study suggested that depletion of the stroma would allow for more effective deliver drugs to the tumor. However, subsequent studies have revealed that the stroma also plays an important role in restraining the growth of the tumor (23,30,32). Depletion of cancer associated fibroblasts in GEM models of PDAC result in poorly differentiated tumors that are more aggressive and less responsive to chemotherapy. Thus, the tumor stroma of PDAC presents a conundrum; its presence enhances the chemoresistance of the cancer cells but its absence results in a more aggressive disease. Perhaps methods that more modestly reduce the tumor stoma, as demonstrated here with BET bromodomain inhibition, may shift the balance towards a favorable therapeutic outcome. Additionally, therapies that involve the depletion of the tumor stroma may need to be combined with those that are effective at treating more aggressive, poorly differentiated tumors. BET bromodomain inhibition may be one such therapy, as it was highly effective in treating poorly differentiated and moderate to poorly differentiated xenograft and GEM model of PDAC employed in our studies.
The contribution of BET proteins has been extensively characterized in hematologic malignancies, and there is now an emerging group of solid tumors that have been described to be dependent on BET bromodomain activity (19,33–43). Our experiments reveal a novel role for BET proteins in regulating SHH secretion and GLI transcriptional activity, defining this family as epigenetic regulators of the SHH-GLI signaling pathway. The in vitro and in vivo efficacy of BET inhibition suggests a clinical opportunity to effectively treat PDAC patients with BET inhibitors.
Supplementary Material
Acknowledgments
Financial Support: This work was supported by a grant from The Andrew L. Warshaw, MD, Institute for Pancreatic Cancer Research (awarded to A.S. Liss) and a grant from the NIH: P01CA117969 (awarded to S.P. Thayer).
We thank N. Bardeesy for his critical reading of this manuscript and Y. Leblanc, N. Valsangkar, G. DiNatale, B. Kulemann, and M. Hewitt for their technical assistance.
Footnotes
Conflict of Interest Disclosure: The following authors are employees of Constellation Pharmaceuticals: Jennifer A. Mertz, Barbara M. Bryant, Andrew R. Conery, Charlie Hatton, and Robert J. Sims III.
References
- 1.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013;63:11–30. doi: 10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]
- 2.Neesse A, Krug S, Gress TM, Tuveson DA, Michl P. Emerging concepts in pancreatic cancer medicine: targeting the tumor stroma. Onco Targets Ther. 2013;7:33–43. doi: 10.2147/OTT.S38111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kopp JL, von Figura G, Mayes E, Liu F-F, Dubois CL, Morris JP, et al. Identification of Sox9-dependent acinar-to-ductal reprogramming as the principal mechanism for initiation of pancreatic ductal adenocarcinoma. Cancer Cell. 2012;22:737–50. doi: 10.1016/j.ccr.2012.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Iacobuzio-Donahue CA, Velculescu VE, Wolfgang CL, Hruban RH. Genetic basis of pancreas cancer development and progression: insights from whole-exome and whole-genome sequencing. Clin Cancer Res. 2012;18:4257–65. doi: 10.1158/1078-0432.CCR-12-0315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jones S, Zhang X, Parsons DW, Lin JC-H, Leary RJ, Angenendt P, et al. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science. 2008;321:1801–6. doi: 10.1126/science.1164368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Biankin AV, Waddell N, Kassahn KS, Gingras M-C, Muthuswamy LB, Johns AL, et al. Pancreatic cancer genomes reveal aberrations in axon guidance pathway genes. Nature. Nature Publishing Group, a division of Macmillan Publishers Limited. All Rights Reserved. 2012;491:399–405. doi: 10.1038/nature11547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sawan C, Herceg Z. Histone modifications and cancer. Adv Genet. 2010;70:57–85. doi: 10.1016/B978-0-12-380866-0.60003-4. [DOI] [PubMed] [Google Scholar]
- 8.Bardeesy N, Aguirre AJ, Chu GC, Cheng K-H, Lopez LV, Hezel AF, et al. Both p16(Ink4a) and the p19(Arf)-p53 pathway constrain progression of pancreatic adenocarcinoma in the mouse. Proc Natl Acad Sci USA. 2006;103:5947–52. doi: 10.1073/pnas.0601273103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hingorani SR, Petricoin EF, Maitra A, Rajapakse V, King C, Jacobetz MA, et al. Preinvasive and invasive ductal pancreatic cancer and its early detection in the mouse. Cancer Cell. 2003;4:437–50. doi: 10.1016/s1535-6108(03)00309-x. [DOI] [PubMed] [Google Scholar]
- 10.Jackson EL, Willis N, Mercer K, Bronson RT, Crowley D, Montoya R, et al. Analysis of lung tumor initiation and progression using conditional expression of oncogenic K-ras. Genes Dev. 2001;15:3243–8. doi: 10.1101/gad.943001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jonkers J, Meuwissen R, van der Gulden H, Peterse H, van der Valk M, Berns A. Synergistic tumor suppressor activity of BRCA2 and p53 in a conditional mouse model for breast cancer. Nat Genet. 2001;29:418–25. doi: 10.1038/ng747. [DOI] [PubMed] [Google Scholar]
- 12.Ting DT, Lipson D, Paul S, Brannigan BW, Akhavanfard S, Coffman EJ, et al. Aberrant overexpression of satellite repeats in pancreatic and other epithelial cancers. Science. 2011;331:593–6. doi: 10.1126/science.1200801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lisovsky M, Dresser K, Woda B, Mino-Kenudson M. Immunohistochemistry for cell polarity protein lethal giant larvae 2 differentiates pancreatic intraepithelial neoplasia-3 and ductal adenocarcinoma of the pancreas from lower-grade pancreatic intraepithelial neoplasias. Hum Pathol. 2010;41:902–9. doi: 10.1016/j.humpath.2009.12.004. [DOI] [PubMed] [Google Scholar]
- 14.Devaiah BN, Lewis BA, Cherman N, Hewitt MC, Albrecht BK, Robey PG, et al. BRD4 is an atypical kinase that phosphorylates serine2 of the RNA polymerase II carboxy-terminal domain. Proc Natl Acad Sci U S A. 2012;109:6927–32. doi: 10.1073/pnas.1120422109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sahai V, Kumar K, Knab LM, Chow CR, Raza SS, Bentrem DJ, et al. BET Bromodomain Inhibitors Block Growth of Pancreatic Cancer Cells in Three-Dimensional Collagen. Mol Cancer Ther. 2014;13:1907–17. doi: 10.1158/1535-7163.MCT-13-0925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Garcia PL, Miller AL, Kreitzburg KM, Council LN, Gamblin TL, Christein JD, et al. Oncogene. Nature Publishing Group; 2015. The BET bromodomain inhibitor JQ1 suppresses growth of pancreatic ductal adenocarcinoma in patient-derived xenograft models. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nolan-Stevaux O, Lau J, Truitt ML, Chu GC, Hebrok M, Fernández-Zapico ME, et al. GLI1 is regulated through Smoothened-independent mechanisms in neoplastic pancreatic ducts and mediates PDAC cell survival and transformation. Genes Dev. 2009;23:24–36. doi: 10.1101/gad.1753809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rajurkar M, De Jesus-Monge WE, Driscoll DR, Appleman VA, Huang H, Cotton JL, et al. The activity of Gli transcription factors is essential for Kras-induced pancreatic tumorigenesis. Proc Natl Acad Sci U S A. 2012;109:E1038–47. doi: 10.1073/pnas.1114168109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dawson MA, Prinjha RK, Dittmann A, Giotopoulos G, Bantscheff M, Chan W-I, et al. Inhibition of BET recruitment to chromatin as an effective treatment for MLL-fusion leukaemia. Nature. 2011;478:529–33. doi: 10.1038/nature10509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Canettieri G, Di Marcotullio L, Greco A, Coni S, Antonucci L, Infante P, et al. Histone deacetylase and Cullin3-REN(KCTD11) ubiquitin ligase interplay regulates Hedgehog signalling through Gli acetylation. Nat Cell Biol. 2010;12:132–42. doi: 10.1038/ncb2013. [DOI] [PubMed] [Google Scholar]
- 21.Coni S, Antonucci L, D’Amico D, Di Magno L, Infante P, De Smaele E, et al. Gli2 acetylation at lysine 757 regulates hedgehog-dependent transcriptional output by preventing its promoter occupancy. PLoS One. 2013;8:e65718. doi: 10.1371/journal.pone.0065718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Long J, Li B, Rodriguez-Blanco J, Pastori C, Volmar C-H, Wahlestedt C, et al. The BET bromodomain inhibitor I-BET151 acts downstream of Smoothened to abrogate the growth of Hedgehog driven cancers. J Biol Chem. 2014 doi: 10.1074/jbc.M114.595348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ozdemir BC, Pentcheva-Hoang T, Carstens JL, Zheng X, Wu C-C, Simpson TR, et al. Depletion of carcinoma-associated fibroblasts and fibrosis induces immunosuppression and accelerates pancreas cancer with reduced survival. Cancer Cell. 2014;25:719–34. doi: 10.1016/j.ccr.2014.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tian H, Callahan CA, DuPree KJ, Darbonne WC, Ahn CP, Scales SJ, et al. Hedgehog signaling is restricted to the stromal compartment during pancreatic carcinogenesis. Proc Natl Acad Sci U S A. 2009;106:4254–9. doi: 10.1073/pnas.0813203106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ji Z, Mei FC, Xie J, Cheng X. Oncogenic KRAS activates hedgehog signaling pathway in pancreatic cancer cells. J Biol Chem. 2007;282:14048–55. doi: 10.1074/jbc.M611089200. [DOI] [PubMed] [Google Scholar]
- 26.Bailey JM, Swanson BJ, Hamada T, Eggers JP, Singh PK, Caffery T, et al. Sonic hedgehog promotes desmoplasia in pancreatic cancer. Clin Cancer Res. 2008;14:5995–6004. doi: 10.1158/1078-0432.CCR-08-0291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thayer SP, di Magliano MP, Heiser PW, Nielsen CM, Roberts DJ, Lauwers GY, et al. Hedgehog is an early and late mediator of pancreatic cancer tumorigenesis. Nature. 2003;425:851–6. doi: 10.1038/nature02009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Collins MA, Bednar F, Zhang Y, Brisset J-C, Galbán S, Galbán CJ, et al. Oncogenic Kras is required for both the initiation and maintenance of pancreatic cancer in mice. J Clin Invest. 2012;122:639–53. doi: 10.1172/JCI59227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Clark CE, Hingorani SR, Mick R, Combs C, Tuveson DA, Vonderheide RH. Dynamics of the immune reaction to pancreatic cancer from inception to invasion. Cancer Res. 2007;67:9518–27. doi: 10.1158/0008-5472.CAN-07-0175. [DOI] [PubMed] [Google Scholar]
- 30.Rhim AD, Oberstein PE, Thomas DH, Mirek ET, Palermo CF, Sastra SA, et al. Stromal elements act to restrain, rather than support, pancreatic ductal adenocarcinoma. Cancer Cell. 2014;25:735–47. doi: 10.1016/j.ccr.2014.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Olive KP, Jacobetz MA, Davidson CJ, Gopinathan A, McIntyre D, Honess D, et al. Inhibition of Hedgehog signaling enhances delivery of chemotherapy in a mouse model of pancreatic cancer. Science. 2009;324:1457–61. doi: 10.1126/science.1171362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee JJ, Perera RM, Wang H, Wu D-C, Liu XS, Han S, et al. Stromal response to Hedgehog signaling restrains pancreatic cancer progression. Proc Natl Acad Sci U S A. 2014;111:E3091–100. doi: 10.1073/pnas.1411679111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Delmore JE, Issa GC, Lemieux ME, Rahl PB, Shi J, Jacobs HM, et al. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell. 2011;146:904–17. doi: 10.1016/j.cell.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mertz JA, Conery AR, Bryant BM, Sandy P, Balasubramanian S, Mele DA, et al. Targeting MYC dependence in cancer by inhibiting BET bromodomains. Proc Natl Acad Sci U S A. 2011;108:16669–74. doi: 10.1073/pnas.1108190108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zuber J, Shi J, Wang E, Rappaport AR, Herrmann H, Sison EA, et al. RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature. 2011;478:524–8. doi: 10.1038/nature10334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Filippakopoulos P, Qi J, Picaud S, Shen Y, Smith WB, Fedorov O, et al. Selective inhibition of BET bromodomains. Nature. 2010;468:1067–73. doi: 10.1038/nature09504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cheng Z, Gong Y, Ma Y, Lu K, Lu X, Pierce LA, et al. Inhibition of BET bromodomain targets genetically diverse glioblastoma. Clin Cancer Res. 2013;19:1748–59. doi: 10.1158/1078-0432.CCR-12-3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shimamura T, Chen Z, Soucheray M, Carretero J, Kikuchi E, Tchaicha JH, et al. Efficacy of BET Bromodomain Inhibition in Kras-Mutant Non-Small Cell Lung Cancer. Clin Cancer Res. 2013;19:6183–92. doi: 10.1158/1078-0432.CCR-12-3904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Puissant A, Frumm SM, Alexe G, Bassil CF, Qi J, Chanthery YH, et al. Targeting MYCN in neuroblastoma by BET bromodomain inhibition. Cancer Discov. 2013;3:308–23. doi: 10.1158/2159-8290.CD-12-0418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Segura MF, Fontanals-Cirera B, Gaziel-Sovran A, Guijarro MV, Hanniford D, Zhang G, et al. BRD4 sustains proliferation and represents a new target for epigenetic therapy in melanoma. Cancer Res. 2013;73:6264–76. doi: 10.1158/0008-5472.CAN-13-0122-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wyce A, Degenhardt Y, Bai Y, Le B, Korenchuk S, Crouthame M-C, et al. Inhibition of BET bromodomain proteins as a therapeutic approach in prostate cancer. Oncotarget. 2013;4:2419–29. doi: 10.18632/oncotarget.1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Asangani IA, Dommeti VL, Wang X, Malik R, Cieslik M, Yang R, et al. Therapeutic targeting of BET bromodomain proteins in castration-resistant prostate cancer. Nature. 2014 doi: 10.1038/nature13229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bandopadhayay P, Bergthold G, Nguyen B, Schubert S, Gholamin S, Tang Y, et al. BET bromodomain inhibition of MYC-amplified medulloblastoma. Clin Cancer Res. 2014;20:912–25. doi: 10.1158/1078-0432.CCR-13-2281. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.