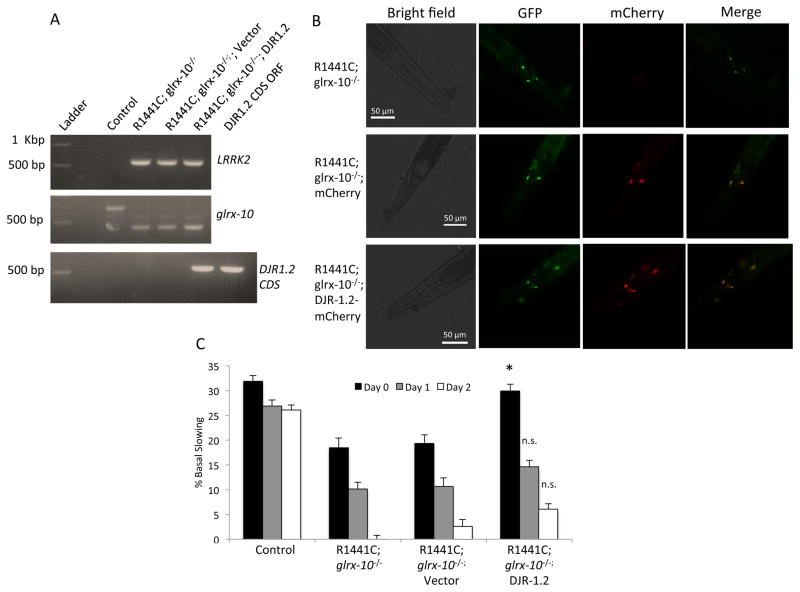
Figure 6. Over-expression of DJR-1.2 partially compensates for the loss of GLRX-10.
(A) PCR analysis of LRRK2 (upper panel), WT or mutant glrx-10 DNA (middle panel), and coding DNA sequence (CDS) of djr-1.2 (lower panel) indicating the generation of R1441C; glrx-10−/−; vector (mCherry) and R1441C; glrx-10−/−; DJR-1.2-mCherry expressing C. elegans lines. (B) Fluorescent images of the head region of the newly generated C. elegans depicting the expression of Pdat-1::mCherry and Pdat-1::DJR-1.2:mCherry in the dopaminergic neurons. Expression of GFP tagged dopaminergic neurons co-localize with expression of both Pdat-1::mCherry and Pdat-1::DJR-1.2::mCherry indicating dopaminergic expression of mCherry and DJR-1.2-mCherry, respectively. (C) Basal slowing data demonstrating that over-expression of DJR-1.2 partially recues the LRRK2-R1441C mediated dysfunction of dopamine-dependent movement in the glrx-10−/− background. *p<0.05 compared to R1441C; glrx-10−/−; vector. Error bars represent SEM. n = 3 independent groups of worms for all days, 10 worms per genotype per experiment. Statistical analysis was completed using one way ANOVA followed by Tukey’s post-hoc test.