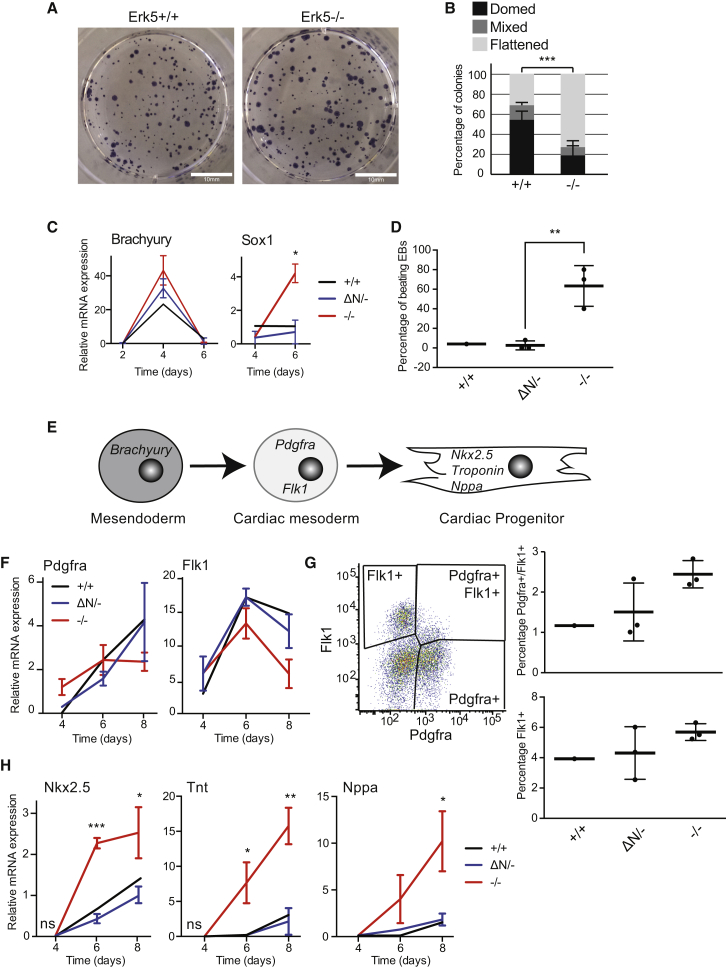
Figure 4.
Erk5 Controls Neuroectoderm and Cardiomyocyte Specification of Differentiating ESCs
(A) Alkaline phosphatase staining of Erk5+/+ and Erk5−/− mESC colonies.
(B) Analysis of colony morphology of Erk5+/+ and Erk5−/− mESC colonies. Data represent average ± SD (n = 3).
(C) Relative mRNA expression of Brachyury and Sox1 were determined for Erk5+/+, Erk5ΔN/− (three independent clones) and Erk5−/− (three independent clones) mESCs. Data represent the average of all clones ± SD from a representative experiment (n = 3).
(D) Percentage of EBs displaying beating areas derived from Erk5+/+, Erk5ΔN/−, and Erk5−/− mESCs. Data represent the average of all clones ± SD from a representative experiment (n = 3).
(E) Scheme outlining stages of cardiac differentiation.
(F) Relative mRNA expression of Pdgfra and Flk1 was determined for Erk5+/+, Erk5ΔN/−, and Erk5−/− mESCs. Data represent the average of all clones ± SD from a representative experiment (n = 3).
(G) FACS quantification of Pdgfra+/Flk1+ cardiac and Flk1+ endothelial progenitors recovered from each cell line. A representative FACS plot illustrating the distinct populations is provided. Data represent average ± SD from a representative experiment (n = 3).
(H) mRNA expression levels of Nkx2.5, Tnt, and Nppa were determined for Erk5+/+, Erk5ΔN/−, and Erk5−/− mESCs. Data represent the average of all clones ± SD from a representative experiment (n = 2).
See also Figure S3.