Figure 1.
Separating Expression of X-Linked Genes into Stromal and Tumor Compartments
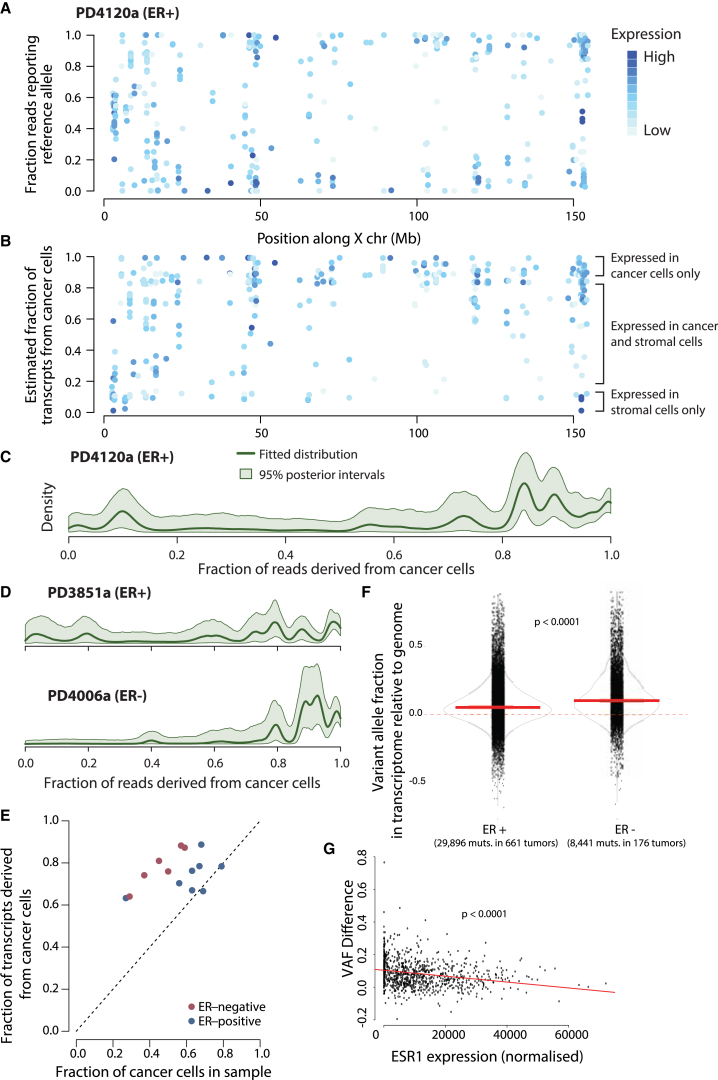
(A) Fraction of RNA-seq reads reporting reference allele of heterozygous germline SNPs on the X chromosome in one of the patients (PD4120a). The depth of color reflects the level of expression.
(B) Fraction of transcripts derived from tumor cells for each heterozygous germline SNP shown in (A), estimated with a Bayesian Dirichlet process, is shown.
(C) Estimated distribution and 95% posterior intervals for relative gene expression in cancer versus stromal cells for PD4120a. The y axis reports the estimated density of genes; the x axis reports the fraction of transcripts for each gene deriving from cancer cells. Thus, the transcripts for most genes in PD4120a are 80%–100% derived from cancer cells and 0%–20% from stromal cells, with only a small peak of genes predominantly expressed from stromal cells.
(D) Distributions for several selected primary cancers are shown, as for (C).
(E) Overall fraction of transcripts derived from cancer cells (y axis) compared to the estimated proportion on tumor cells in the sample (x axis, estimated from genomic DNA using copy-number profiles) is shown.
(F) Increased expression of the mutated allele in ER− as compared to ER+ breast cancer transcriptomes (plotted relative to the genome). Primary breast cancers sequenced as part of TCGA are shown. Plotted on the y axis is the variant allele fraction in the transcriptome, relative to the genome (VAFdiff).
(G) Inverse relationship between each tumors’ expression of Estrogen Receptor 1 (ESR1) and the overall expression of its point mutations (shown as VAFdiff; −0.2433, p < 0.0001). Using linear regression analysis to model this relationship, we determined that, for every 1% drop in ESR1, ∼15 additional point mutations are expressed.