FIGURE 1.
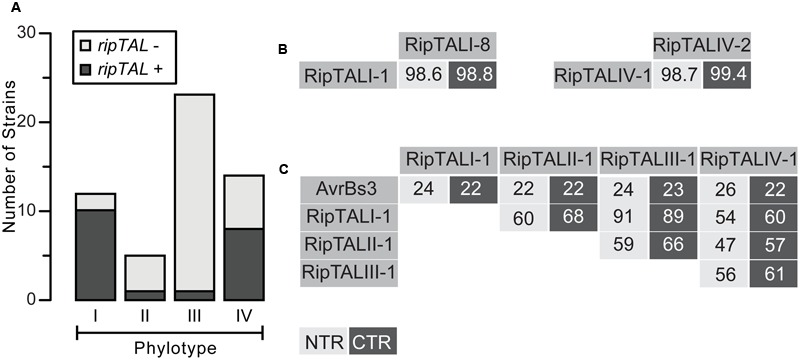
RipTAL abundance differs across Ralstonia solanacearum phylotypes but RipTALs sequences are similar within and different across phylotypes. (A) Abundance of ripTALs in strains from distinct phylotypes. The assessment is based on PCR analysis with primers flanking the repeats and was carried out on a broad collection of Rssc strains covering all phylotypes and different degrees of host adaptation. (B,C) Pairwise NTR and CTR sequence identities of depicted RipTALs from closely related Rssc strains (B) and RipTALs from different Rssc phylotypes or the Xanthomonas TALE AvrBs3 (C) are given in percent. Light and dark gray background indicates identities in the NTR and CTR, respectively.
