FIGURE 2.
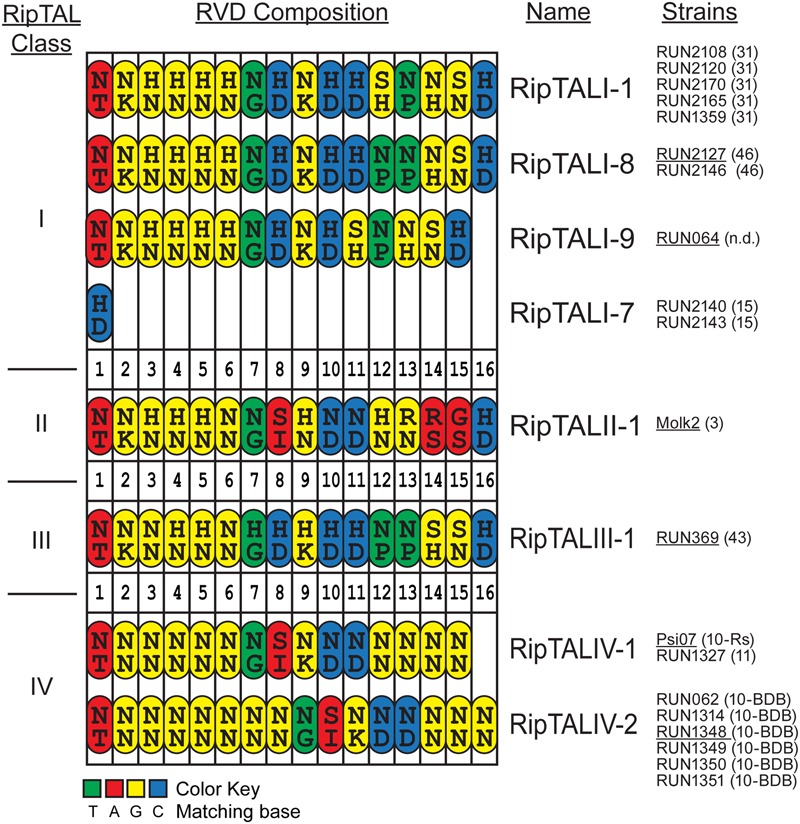
Comparison of RVD compositions of novel RipTALs across all four Ralstonia solanacearum phylotypes reveals limited diversity. Cartoon displays RVD compositions of newly identified RipTALs separated by class. Each repeat is depicted as an oval. Capital letters inside the repeats indicate amino acids (single letter code) in position 12 and 13 (RVD) of each repeat. Repeats are color-coded based on the preferred base of repeat residue 13, which is the key base specificity determinant, with a color code given at the bottom. Strains bearing a particular ripTAL are given next to the RipTAL identifier in black text. Text in brackets gives the sequevar of this strain, n.d. indicates that this strain has not been clearly assigned to a sequevar. Underlined strain name indicates that the given ripTAL was studied as a representative in functional assays. RipTALI-2 to RipTALI-6 were described previously (de Lange et al., 2013) and are shown in Supplementary Figure S1.
