FIGURE 9.
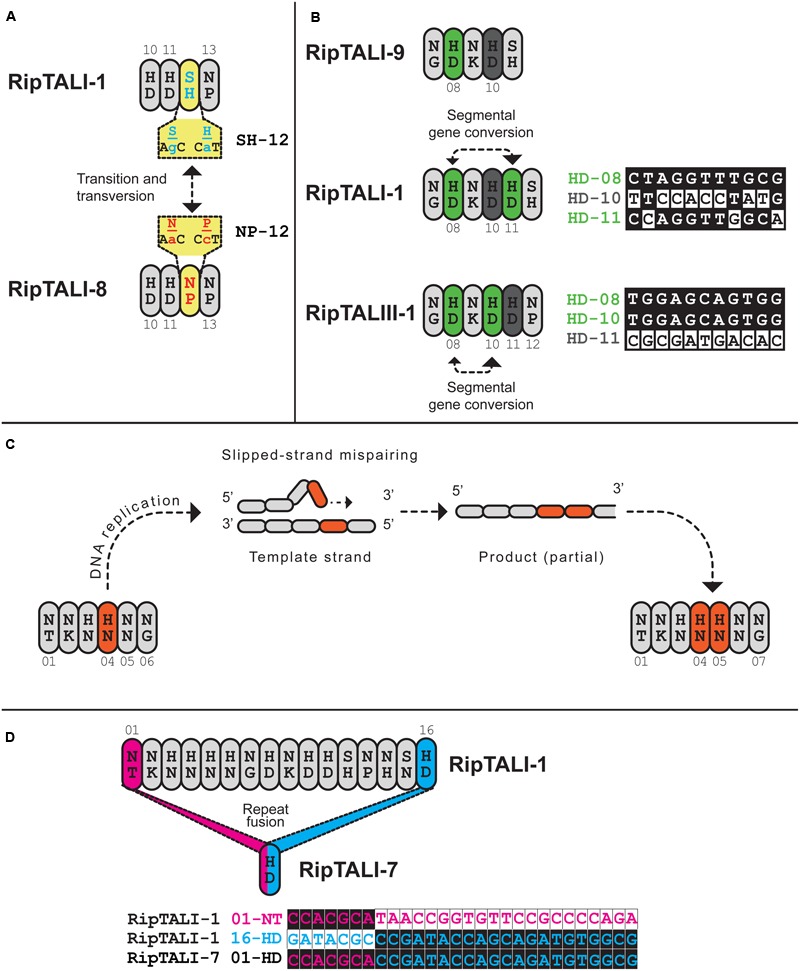
Inspection of ripTAL CRDs indicates the molecular mechanisms shaping ripTAL CRD composition. Closely related repeats are colored in the same color, with exception of gray that does not indicate any relatedness. (A) Specificity altering SNPs. Repeats 10-13 of RipTALI-1 and RipTALI-8 are shown. The RVDs and cognate codons of repeat 12 of each RipTAL are further given in yellow boxes. The two depicted SNPs constitute the only polymorphisms between these two repeats. (B) Repeat duplication by segmental gene conversion. Repeats 7–11 of RipTALI-9 are shown, as well as repeats 7-12 of RipTALI-1 and RipTALIII-1. Green color indicates HD repeats that are highly similar in sequence, within each array. A less related HD repeat is displayed in dark gray. Proposed segmental gene conversion events are indicated by dashed lines with arrowheads. Polymorphic bases between repeats 8, 10, and 11 of the respective ripTAL are displayed to the right of the cartoon display. Next to the base comparison, repeat RVD, as well as the position within the array, colored according to the fill color of that repeat in the cartoon display is given. (C) Duplication of adjacent repeats by slipped-strand mispairing exemplified on ripTALII-1. Repeats 1-6 of a proposed ancestral repeat array are shown to the left. A slipped-strand mispairing DNA intermediate is shown above. Repeats 1–7 of the resulting product are shown to the right. Slipped-strand mispairing leads to a duplication of the repeat colored in orange. (D) A recombination event leads to loss of all repeats except one in ripTALI-7. The remaining repeat is fusion of ripTALI-1 repeats 1 (pink) and 16 (blue). To the right, polymorphic bases of repeats 1 and 16 are shown. The ripTAL designation, repeat RVD and repeat position are given to the right of the sequence of polymorphic bases, colored according to the fill color of that repeat.
