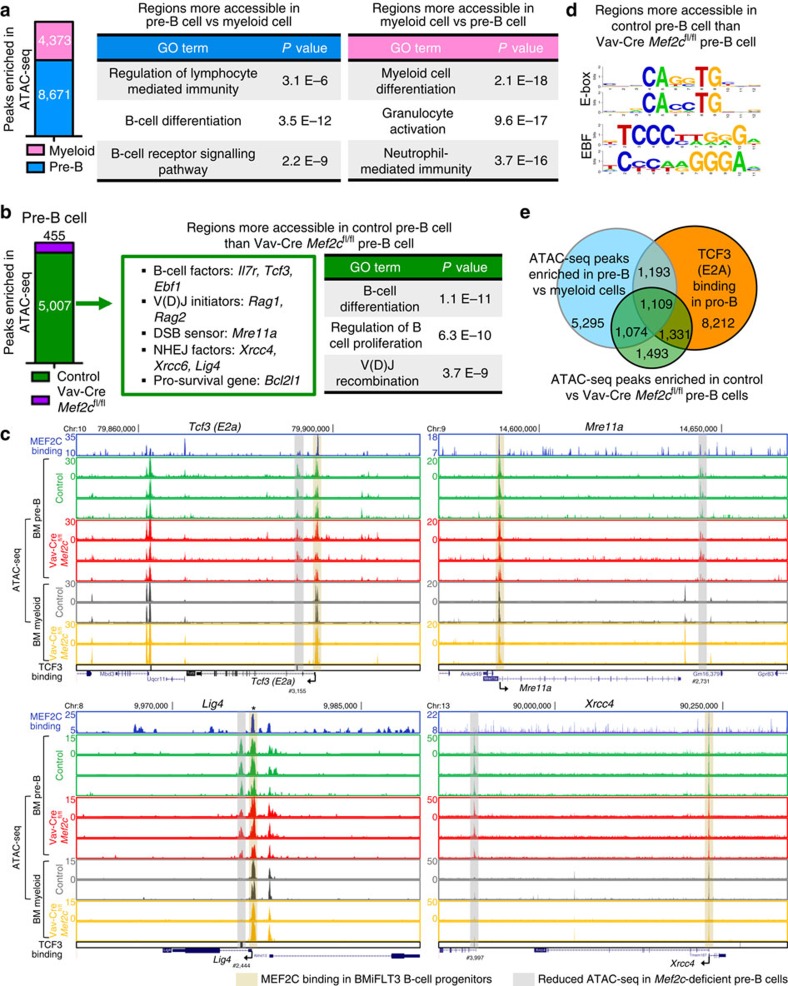
Figure 7. MEF2C promotes chromatin accessibility of its target genes in mouse pre-B cells.
(a) MAnorm analysis of ATAC-sequencing(ATAC-seq) data in control pre-B cells and control bone marrow myeloid (Mac1+Gr1+) revealed that the accessibility of regulatory sites in lineage-specific genes is differentially (ratio≥4, P value≤0.01) regulated in these two populations. (b) MAnorm analysis of ATAC-seq data in control and Mef2c-deficient pre-B cells revealed that MEF2C is required to promote chromatin accessibility in regulatory regions of genes required for B-cell differentiation. (c) Loss of Mef2c reduced the chromatin accessibility nearby genes that are critical for B-cell differentiation and DNA repair. Many of these regulatory sites are also uniquely accessible in pre-B cells as compared with myeloid cells. Regions that are significantly (ratio≥4, P value≤0.01) more accessible in control than Mef2c-deficient pre-B cells are highlighted in grey and labelled with reference numbers co-related to Supplementary Data 4. Regions that are bound by MEF2C are highlighted in brown. MEF2C bindings that do not yield statistical significance are labelled with asterisks. (d) The ATAC-seq peaks that are more accessible in control pre-B cells compared with Mef2c-deficient pre-B cells are enriched for E-box and EBF motifs. (e) Venn diagram showing extensive overlap of regions that are more accessible in control pre-B cells vs. Mef2c-deficient pre-B cells with regions that are more accessible in pre-B cells than myeloid cells, as well as TCF3-bound regions in pro-B cells.