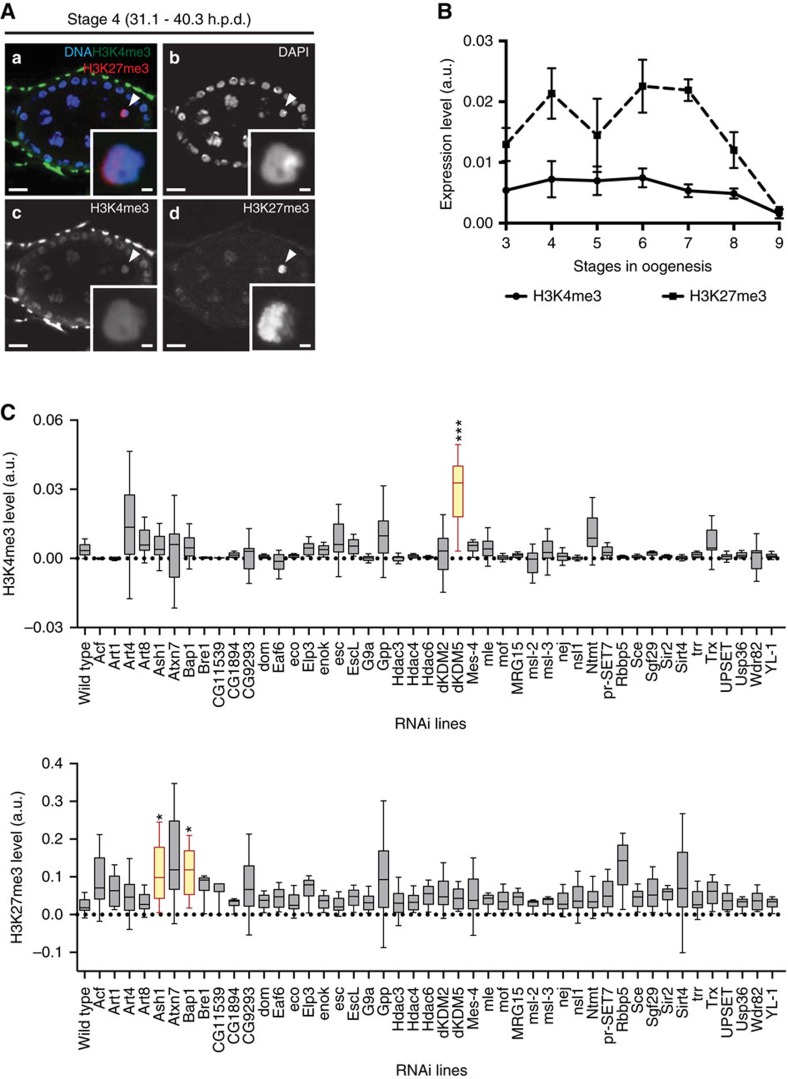
Figure 2. A germ line-specific in vivo RNAi screen identified three major regulators of the oocyte epigenome.
(A) The Drosophila oocyte has both activating (histone H3 lysine 4 trimethylation—H3K4me3) and repressive (histone H3 lysine 27 trimethylation—H3K27me3) marks (a–d). Development time in relation to the start of oogenesis is expressed in hours post-germ line stem cell division (h.p.d.). Arrowheads and insets indicate oocyte chromatin. Scale bars, 10 μm for ovarian follicles and 1 μm for oocyte insets. (B) Temporal analysis of oocyte H3K27me3 and H3K4me3 levels throughout oogenesis. The high levels of H3K27me3 during the prophase I arrest are marked by a reduction prior to the onset of oocyte transcriptional reactivation (stage 9). H3K4me3 levels are kept at a low level throughout prophase I. Histone post-translation modifications (PTMs) relative levels are expressed in fluorescence arbitrary units (a.u.). Error bars represent the s.e.m. A total of six replicates were analysed for each data point. (C) Effects of the RNAi-mediated knockdown of different chromatin remodellers on oocyte H3K4me3 (top) and H3K27me3 (bottom) levels. A total of 46 different chromatin remodellers were compatible with the development to mid-oogenesis when depleted specifically in the germ line. Of these, three (dKDM5, Ash1 and Bap1) introduced significant disturbances to the oocyte epigenome. Relative histone PTM signals are expressed in fluorescence arbitrary units (a.u.), horizontal lines specify median values, error bars represent s.d. and asterisks indicate significant difference (paired t-test; P<0.04).