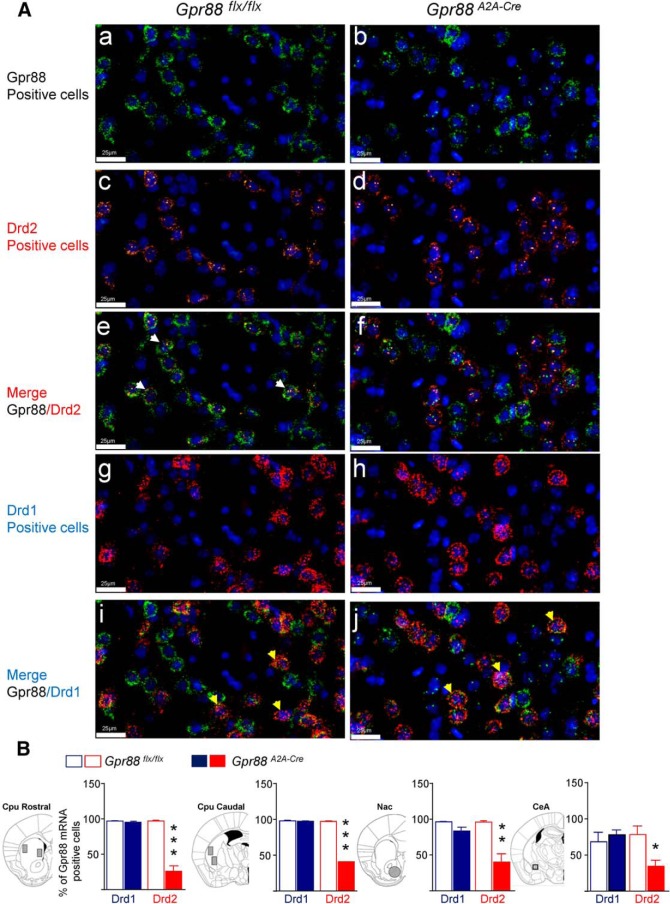
Figure 1.
Molecular characterization of conditional A2AR-Gpr88 KO mice. A, Triple-fluorescent in situ hybridization probing Gpr88 (Aa, Ab, Ae, Af, Ai, and Aj; probe labeled in green), Drd2 (Ac–Af; probe labeled in orange), and Drd1 (Ag–Aj; probe labeled in red). Representative images are shown. In Gpr88flx/flx control animals, Gpr88 mRNA colocalizes with both Drd2 (Ae: merge GPR88/Drd2, white arrows) and Drd1 mRNA (Ai: merge GPR88/Drd1, yellow arrows). In contrast, Gpr88A2A-Cre conditional mice show almost no colocalization with Drd2 (Af: merge GPR88/D2R), while colocalization with Drd1 remains (Aj: merge GPR88/Drd1, yellow arrows). DAPI staining (blue) was used to label all cell nuclei. Scale bar, 25 µm. B, Quantification shows a strong decrease of Gpr88/Drd2 double-positive neurons (red), but not GPR88/Drd1 double-positive (blue) neurons, in the CPu, Nac, and CeA of Gpr88A2A-Cre conditional mice. Percentage of Gpr88 expression was calculated based on the total number of Drd1- or Drd2-positive cells counted [(number Drd1- or Drd2-expressing cells coexpressing Gpr88 × 100)/total number of Drd1- or Drd2-expressing cells]. Data are presented as the mean ± SEM. n = 4, Gpr88A2A-Cre; n = 4, Gpr88flx/flx; solid asterisks). *p < 0.05; **p < 0.01; ***p < 0.001 (Student’s t test).