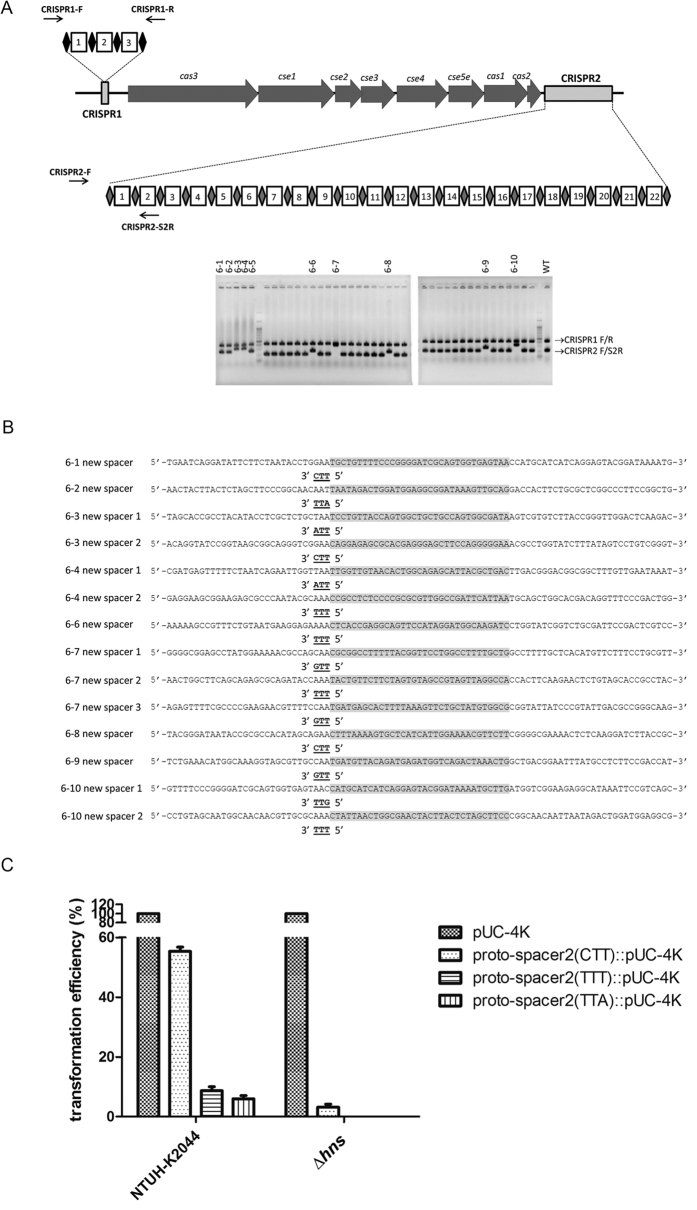
Figure 4. Acquisition of new spacers and PAM sequences in K. pneumoniae.
The acquisitions of new spacers into CRISPR1 and CRISPR2 arrays were detected by PCR simultaneously using primers (CRISPR1-F and CRISPR1-R for CRISPR1; CRISPR2-F and CRISPR2-S2R for CRISPR2) indicated by the arrows (Fig. 4A). Ten ∆hns clones (6-1~6-10) loss of pUC-4K plasmid were detected to have elongated CRISPR2, whereas wild type strain served as a control. Fourteen proto-spacers (marked in gray) and adjacent sequences (upstream and downstream 30 bps) on pUC-4K plasmid were aligned (Fig. 4B). The possible PAM sequences were showed in bold and underlined. The transformation efficiencies of pUC-4K and pUC-4K with engineered proto-spacer 2 carrying different PAM sequences were compared in NTUH-K2044 wild type and hns deletion (∆hns) (Fig. 4C). The transformation efficiency of pUC-4K was set as 100% and those of proto-spacer2::pUC-4K with different PAM sequences were calculated accordingly. Data are presented as means ± SEM from three independent experiments.