Fig. 3.
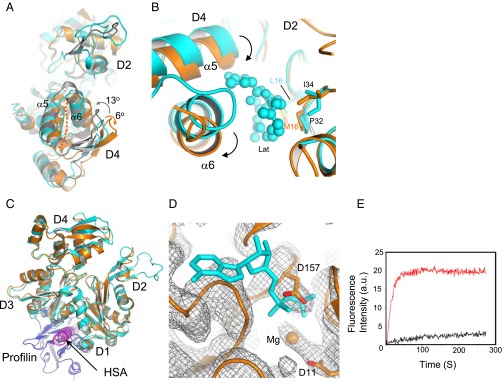
Structural features of N-actin complexed with Arp4. (A) Superposition of N-actin (orange), G-actin [cyan; Protein Data Bank (PDB) ID code 1YAG], and an actin protomer in F-actin (gray, PDB ID code 3MFP). The structural alignments were constructed on the major domain D1–D2. Upon polymerization, the two major domains of G-actin show a counterclockwise rotation of ∼13° and become flat in F-actin, whereas it is additionally twisted by a clockwise rotation of ∼6° in the N-actin complex. (B) Conformational changes in the major-domain cleft of N-actin around the latrunculin-binding pocket. N-actin was structurally aligned with G-actin (PDB ID code 1ESV). Latrunculin (Lat), in a ball-and-stick model, is surrounded by hydrophobic contacts from L16, P32, and I34 in D2 subdomain of G-actin. The arrows indicated the movement of α5 and α6. (C) Superposition of the structures of N-actin and G-actin bound to profilin (blue; PDB ID code 2BTF). The HSA helix bound to the hydrophobic cleft of N-actin clashes with profilin. (D) Superposition of the structure of N-actin with the electron density map (contour level σ = 1) of the nucleotide-binding pocket. The ADP bound by G-actin (PDB ID code 1YAG) is indicated with a stick model. No electron density at the position of ADP was found in the crystal structure of the N-actin complex. (E) Time-course of the nucleotide exchange by G-actin (red curve) and the N-actin complex (black curve). Conditions: 3 μM G-actin or N-actin complex was mixed with 50 μM ε-ATP in buffer (2 mM Tris⋅HCl, pH 7.5 and 50 mM KCl).