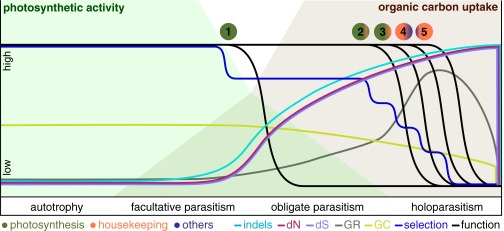
Fig. 3.
General model of plastome evolution under relaxed functional constraints. The model illustrates the evolution of genomic traits (GC, GC content; GR, genomic changes including gene deletions), functionality of different plastid-encoded complexes (blue, other metabolic genes; green, photosynthesis genes; orange, housekeeping genes), evolutionary rates (dN, dS) as well as selection intensity and their associations during the transition from autotrophy with high photosynthetic activity (green background) and no uptake of external organic carbon to holoparasitism with maximal uptake (brown background). As the specialization on the heterotrophic lifestyle increases, more plastid-encoded complexes are lost, altering evolutionary rates and the genome structure. Numbers indicate the predicted functional reductions starting with the loss of nonessential or stress-relevant genes (ndh genes) (1) followed by primary photosynthesis genes (pet and psa/psb genes) and the plastid-encoded polymerase (PEP) (2), genes that have a prolonged function (e.g., atp genes, rbcL) and nonessential housekeeping genes (3), other metabolic genes (e.g., accD, clpP, ycf1/2) (4), and, finally, the remaining housekeeping genes, including trnE (5). Unlike most factors that are likely to show a rather monotonic trend as parasitism evolves, selectional strength may experience several periods of relaxation and intensification. The associations of evolutionary rates, genomic traits, and functional losses between 1 and 3 have been observed, whereas those after 3 are hypothetical. For simplicity, we here depict the functional reduction of a plastome with a coding capacity of flowering plants; more or fewer lineage-specific reconfigurations may occur in other lineages.