Abstract
Mitochondrial DNA (mtDNA) profiles can be classified into phylogenetic clusters (haplogroups), which is of great relevance for evolutionary, forensic and medical genetics. With the extensive growth of the underlying phylogenetic tree summarizing the published mtDNA sequences, the manual process of haplogroup classification would be too time-consuming. The previously published classification tool HaploGrep provided an automatic way to address this issue. Here, we present the completely updated version HaploGrep 2 offering several advanced features, including a generic rule-based system for immediate quality control (QC). This allows detecting artificial recombinants and missing variants as well as annotating rare and phantom mutations. Furthermore, the handling of high-throughput data in form of VCF files is now directly supported. For data output, several graphical reports are generated in real time, such as a multiple sequence alignment format, a VCF format and extended haplogroup QC reports, all viewable directly within the application. In addition, HaploGrep 2 generates a publication-ready phylogenetic tree of all input samples encoded relative to the revised Cambridge Reference Sequence. Finally, new distance measures and optimizations of the algorithm increase accuracy and speed-up the application. HaploGrep 2 can be accessed freely and without any registration at http://haplogrep.uibk.ac.at.
INTRODUCTION
Mitochondrial DNA (mtDNA) haplogroup classification has been mandatory in the area of phylogenetic and forensic genetics, and it is now also increasingly applied in medical genetics. For haplogroup classification, HaploGrep (1) provides a fully automated way to determine haplogroups by traversing the underlying phylogenetic tree Phylotree (2). Although HaploGrep has been widely accepted and used by a continuously growing group of researchers, important features to support high quality mtDNA research projects with a profound quality control (QC) (3,4), standardized formats or additional features for data analysis were lacking in the first version. While several new haplogroup classification tools were published subsequently (5–9), addressing issues with the reference sequence (10), QC was widely neglected. QC was already fundamental for Sanger based sequencing, and Next Generation Sequencing (NGS) requires even more attention (11–14).
Since the availability of NGS devices have increased significantly during the last years, the processing costs of mtDNA in the laboratory decreased. This resulted in an explosion of newly generated data. Furthermore, the number of future mtDNA NGS sequencing studies is expected to grow continuously. Consortia like the ‘Centers for Common Disease Genomics’ (CCDG) are expected to sequence 150,000 to 200,000 whole genomes within the next four years (http://www.genome.gov/27563453). The 1000 Genomes Project (15) already contributed over 2,500 sequences to the expansion of the phylogenetic knowledge, showing high inter- and intra-population haplogroup diversity (16). Data from such projects highlight also the need for direct data import avoiding manual conversion steps. So far, users had to use mitoSAVE (17) in combination with Microsoft Excel to convert VCF files (18) into HaploGrep's initial input format hsd. Furthermore, the underlying algorithm requires adaptions to account for large datasets containing thousands of samples (19,20), and to account for the growing sequence archive within GenBank, the major source for the underlying phylogenetic tree. While the first release of Phylotree incorporated in HaploGrep was version 10 with 2,192 haplogroups, the current Phylotree version 17 (16) comprises 5,437 haplogroups, refining the human mtDNA tree even further. To detect differences in haplogroup classification results conducted on older or newer versions of Phylotree as well as to reproduce their results, multi-version support of Phylotree is required.
Several well established tools for further mtDNA data analysis such as ARLEQUIN (21), PAUP* (22) or MRBAYES (23) exist, but can currently not deal with the provided export format of HaploGrep. There is a need to support additional output formats in order to eliminate error-prone and time-intensive conversions. Phylogenetic research strives for improving the current knowledge of the worldwide mitochondrial phylogenetic tree by exhibiting important new haplogroups, which is usually accompanied by a tree diagram combining old and new groups. Phylogenetic trees, displaying the relation between a large number of haplotypes, cannot be constructed by hand conveniently, and the manual process is prone to errors. Direct support of generating phylogenetic trees will greatly help to standardize the detection of novel haplogroups.
To address the aforementioned shortcomings, we developed HaploGrep 2. It includes improvements to the current functionality, new input formats and several quality checks. The modular architecture allows us to adapt it to future needs by simple adding rules and new components without altering the core of the HaploGrep classification algorithm.
MATERIALS AND METHODS
HaploGrep 2 is a web application that communicates through a REST API with the web server. Thus, all computation intensive tasks are executed directly on the server. The haplogroup classification itself is based on pre-calculated phylogenetic weights that correspond to the occurrence per position in Phylotree and reflecting the mutational stability of a variant. In the updated classification algorithm, the weights are now scaled from 1 to 10 in a non-linear way (see Supplementary Table S1). Thus, the rare occurrences of variants in Phylotree will no longer influence the classification toward those haplogroups as much as in the previous version. Once the data is imported, the haplogroup classification is started automatically. Optimizations within the code led to a 20-fold speed-up compared to HaploGrep 1. By storing only the 50 highest ranked haplogroups per sample the memory consumption could be reduced significantly.
Furthermore, new dissimilarity metrics for the mtDNA haplogroup classification were introduced. In addition to the already implemented Kulczynski distance (1), the Jaccard index, the Hamming distance and the Kimura 2-parameter distance were included (24) (see Supplementary Table S2 and 3 for performance comparison). Further major improvements included a check for artificial recombination (25) and a check for systematic artefacts and for rare or potential phantom mutations (26). For detecting artificial recombination, we apply two different strategies: the first strategy, proposed by Kong et al. (27), counts the remaining variants that were not assigned to the resulting best haplogroup, and tests whether these variants could be assigned to another haplogroup. For this step, mutational hotspots are excluded (e.g. 315.1C or 16519). The second recombination strategy assumes prior knowledge about the specific placement of the fragments of the polymerase chain reaction products (amplicons). With this information in hand, a check comparing the profiles relative to the fragment ranges can be executed. The user-defined fragments are generated, and the profiles split accordingly. If the distance of both haplogroup fragments exceeds five phylogenetic nodes, the sample is listed as potentially contaminated.
RESULTS
The new HaploGrep 2 web server can be accessed freely without registration at http://haplogrep.uibk.ac.at and works with all current browser versions. In order to simplify the integration within external workflows and pipelines, we further provide a stand-alone version, a command line version, and a Rest API of the web server. This allows simplified integration within external workflows and pipelines, as e.g. already used within the mtDNA-Server (28) (http://mtdna-server.uibk.ac.at) or the MitoMaster project (29,30). Details and further information can be found on the project websites.
Input
HaploGrep 2 supports the direct import of VCF files (18), one of the current standard formats for genetic data. This new import format is implemented by using HTSJDK, a Java API for high-throughput sequencing data formats (http://samtools.github.io/htsjdk/). For validation purposes, we used samples from the 1000 Genomes Project (1000G) Phase 1 and Phase 3. Through VCF support, output files from NGS software like the IonTorrent Suite can now be handled directly within HaploGrep 2. A VCF upload can also be performed by using the new Rest API, resulting in a JSON-formatted string including the haplogroup status and the quality score for each sample.
Quality control using a rule-based engine
The new rule-based engine performs several QCs: (i) check amount of remaining variants, that were not assigned to the resulting best haplogroup, (ii) amount of variants not used for the best ranked haplogroup per sample, (iii) differences between provided and estimated haplogroup, (iv) quality scores for haplogroup assignment, (v) ambiguous haplogroup classification, (vi) check for heteroplasmic variants, which is of major interest in mtDNA NGS studies, (vii) check for ‘N’ nucleobases, indicating sequencing or genotyping problems and (viii) reference sequence problems, triggered by profiles showing explicit variants identical to the rCRS (31) characteristics (e.g. 263A, 1438A, 8860A, etc.). For example, with the help of rule (ii) we could identify problems within the provided 1000G Phase 3 mtDNA data, where variants at prominent positions (e.g. 152, 195, 263) were missing, indicating potential problems when writing adjacent variants to the VCF file. Rule (i) and (viii) helped identifying issues with the reported H2 haplogroup in (32) which was in fact a C4a1a1 haplotype, after remapping the positions to the rCRS. The Genome Reference Consortium initially provided a Yoruba reference sequence (before using the rCRS in GRCh37), which was also a source of errors and gets checked with rule (viii) now. All quality checks generate errors (red) and warnings (yellow) by direct representation within a new ‘Errors & Warnings’ tab. This system allows us to react on new demands by adding new rules to the current rule engine (e.g., nomenclature checks).
Artificial recombination and phantom mutations
The artificial recombinants from forensic databases listed in Bandelt et al. (33), which were not recognized accurately in the initial version of HaploGrep, have now been reassessed within HaploGrep 2. The first check for remaining variants (rule (i)) already flagged six out of the seven cases. This rule check is executed automatically when determining new haplogroups and is included in the rule-engine tab. The range-based recombination strategy (Button ‘Check for Recombination’) successfully found all recombined samples. Therefore HaploGrep 2 marks all cases as potentially recombined.
For rare mutations and potential phantom mutations (26,34) (Button ‘Check for Phantom Mutation’), HaploGrep 2 takes phylogenetic knowledge into account. It incorporates all variants with frequency scores from Soares et al. (35), based on 2,196 complete mitochondrial genomes. All remaining variants in a sample are annotated according to this list. If (i) a variant occurs with a frequency less than three times and (ii) at least two samples share this variant, then it is listed as a rare mutation in the report. The reason for this threshold is that known phantom mutations with score 2 are in fact, also present within the rare mutation list. Therefore this filter allows the identification of potential phantom mutations but also mapping/alignment problems, which will become more prominent when applying mapping algorithms regardless of the phylogeny. Also problems with the correct mtDNA nomenclature are represented in this report (cf. the analysis of 1000G Phase 3 data, where 832 of 2,504 samples showed 3107C whereas 13 showed 3109d, likely triggered by the coding 3107N of the void position 3107 in the rCRS).
Distance concordance check
Besides the Kulczynski distance used for haplogroup estimation in the first HaploGrep version, we added three new dissimilarity indices: Hamming distance, Jaccard index as well as the Kimura 2-parameter (Kimura2P) distance based on transition and transversion rates. For validation purposes we applied all four distances to the 1000G Phase 1 data (n = 1,074) as well as to the dataset provided by Li et al. (19), including 2,000 exome sequencing data from a Danish cohort. Table 1 presents the summary of the distance concordance check. While the runtime of the first three distances was almost identical (4.6–4.7 s), the Kimura2P distance showed a 33-fold higher wall-time (158.1 s). The ratio was similar for the Li data (6.4 s for Kulczynksi versus 210 s for Kimura2P). With different results in 21 out of the 1074 samples (1.96%) in the 1000G Phase 1 data, 153 out of 2534 (6.11%) in the 1000G Phase 3 data and 98 (4.9%) out of 2000 in the exome sequencing samples, the user receives additional information about data quality and can check suspect samples. Employing the distance concordance check, coverage problems in the exome data and problems with the VCF file in the 1000G Phase 3 data become visible (see Table 1 and Supplementary Tables S2–4). We therefore provide this combined haplogroup estimation mode (Button ‘Haplogroup Discordance Check’), which lists all samples where at least one metric results in a different result.
Table 1. HaploGrep 2 runtime and concordance over different metrics.
| NGS mtDNA dataset | Sample size | Full concordance over all metrics | HaploGrep 2 Runtime (including QC) |
|---|---|---|---|
| 1000G Phase 1 | 1,074 | 98.0% | 5.7 s |
| Li et al. | 2,000 | 95.1% | 7.7 s |
| 1000G Phase 3 | 2,504 | 93.9% | 13.0 s |
Full concordance means that all samples are classified into the same haplogroup for the different metrics. The HaploGrep 2 runtime refers to the calculation of the Kulczynski distance including all quality checks from the rule-based engine. The detailed results are provided in Supplementary Tables S2, 3 and 4.
Output formats
HaploGrep 2 displays the classification results and all details directly in the browser. Additionally, it provides several new browser- and file-based outputs:
Haplogroups Report: summarizes the graphical output, comprising the quality score, remaining polymorphisms, polymorphisms not found in the top ranked haplogroup, the corresponding amino acid changes (36) and the input profile.
Multiple Alignment Format: based on the rCRS, a multiple sequence alignment format is generated and can be directly viewed in the browser by using BioJS (37). With the help of this export, the freely available PGDSpider (38) can convert the result into a variety of population genetics formats (e.g. ARLEQUIN (21), MEGA (39), MIGRATE (40), PHYLIP (41) directly or the NEXUS format (42) for MRBAYES (23) or PAUP*(22).
VCF: a column-based representation of all samples. For the graphical representation the metadata in the VCF header is skipped, and only the data lines are presented. The complete VCF file can be downloaded for further analysis such as FST computation, linkage disequilibrium (LD), or Principal Component Analysis (PCA), by applying VCFtools (18) directly or to generate PED and MAP files for further analysis with the toolset PLINK (43).
FASTA: each sample is exported as sequence entry in one summarizing fasta file, excluding the alignment information.
Phylogenetic tree visualization
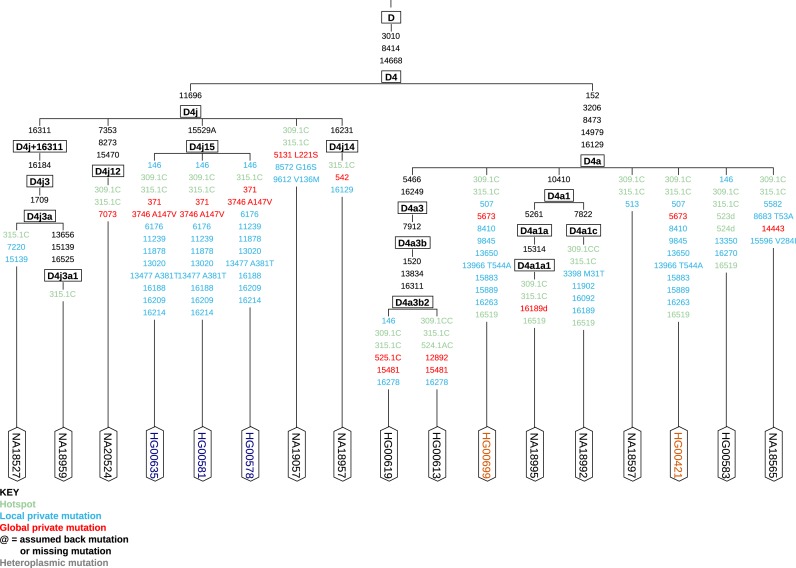
For many researchers a graphical representation of all haplogroups is the most attractive feature, as it shows how new haplogroups fit into the existing Phylotree. This is performed automatically by HaploGrep 2, while all classified samples are combined to a resulting (rooted) tree including all related polymorphisms relative to the rCRS. To customize the output, the user can choose whether hot spot mutations should be taken into account and which export format should be used (see Figure 1 for an example). The tree can be opened directly in the browser or can be downloaded either as pdf, svg or png file. The downloaded pdf and svg file can be further adapted by the user with vector graphic tools like Inkscape (freely available at https://inkscape.org/) or Corel Draw. This output format is implemented by using the Apache Batik SVG Toolkit (https://xmlgraphics.apache.org/batik/).
Figure 1.
Excerpt of the 1000G Phase 1 data generated with the new provided ‘Graphical Phylogenetic Tree’. Polymorphisms in the tips of the phylogeny are candidates for new haplogroups, see for instance the samples belonging to haplogroup D4j15, (confirmed to be related (ftp://ftp.1000genomes.ebi.ac.uk/vol1/ftp/technical/working/20130606_sample_info/20130606_sample_info.xlsx)) or samples HG00699 and HG00421 (not related). Polymorphisms marked in red are not occurring in Phylotree and may require additional attention, whereas mutations in blue are private polymorphisms for this group, already known by Phylotree. The annotation of amino acid changes and mutational hotspots (green) can be defined by the user, thereby hotspots at positions 16182, 16183 and 16519, AC insertion and deletions at 515–524, inserts at 16193 as well as variation around position 310 and point heteroplasmies can be excluded for the phylogenetic reconstruction.
Rest API
Many users are utilizing the command line version of HaploGrep, which is often integrated in workflows. The provision of a Rest API can simplify this, by providing the latest version of HaploGrep 2 to end users. We added a new web resource (haplogrep-ws) to HaploGrep 2 in which VCF files can be uploaded, and the results formatted as a JSON string are exported (see http://haplogrep.uibk.ac.at/blog for an example).
Support of multiple Phylotree versions
Phylotree gets updated constantly. To examine whether an updated version of Phylotree affects previously determined haplogroups, users are now able to select the Phylotree version manually. After changing the version of Phylotree within HaploGrep 2, samples are re-classified automatically and haplogroup updates are listed in brackets, thereby also triggering the rule-based engine and emitting a warning if a sample is classified as belonging to different haplogroups.
DISCUSSION
Several tools similar to HaploGrep have been published since its first release in 2011 (5–9). While haplogroup are estimated by most of these tools with similar performance (33), only a few tools allow for adequate QC. With growing phylogenetic knowledge, mitochondrial haplogroups increasingly gain importance in investigating the correctness of mitochondrial sequences or genotypes, making phylogenetic inference indispensable. Since HaploGrep 2 is based on Phylotree, results are highly dependent on these underlying data, and the results should not be accepted blindly. The growing sample sizes in studies require higher speed while simultaneously maintaining accuracy. We therefore implemented new distance metrics, updated the algorithm, and developed a rule-based engine for additional QC. All these new features can help researchers avoiding inadvertent interpretations of putative hits before publication. HeploGrep 2 will be frequently updated to meet new requirements and demands.
Supplementary Material
Acknowledgments
We thank Luisa Pereira and David Samuels for providing the annotation of the amino acid changes, Matthias Weiler and Bernhard Rupp for helpful comments.
Footnotes
Present address: Dr Sebastian Schönherr, Division of Genetic Epidemiology, Department of Medical Genetics, Molecular and Clinical Pharmacology, Medical University of Innsbruck, Innsbruck 6020, Austria.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
The project was supported by the Austrian Cancer Society/Tirol and by the Tyrolean Science Fund (Tiroler Wissenschaftsfonds).
Conflict of interest statement. None declared.
REFERENCES
- 1.Kloss-Brandstätter A., Pacher D., Schönherr S., Weissensteiner H., Binna R., Specht G., Kronenberg F. HaploGrep: a fast and reliable algorithm for automatic classification of mitochondrial DNA haplogroups. Hum. Mutat. 2011;32:25–32. doi: 10.1002/humu.21382. [DOI] [PubMed] [Google Scholar]
- 2.van Oven M., Kayser M. Updated comprehensive phylogenetic tree of global human mitochondrial DNA variation. Hum. Mutat. 2009;30:E386–E394. doi: 10.1002/humu.20921. [DOI] [PubMed] [Google Scholar]
- 3.Salas A., Carracedo A., Macaulay V., Richards M., Bandelt H.-J. A practical guide to mitochondrial DNA error prevention in clinical, forensic, and population genetics. Biochem. Biophys. Res. Commun. 2005;335:891–899. doi: 10.1016/j.bbrc.2005.07.161. [DOI] [PubMed] [Google Scholar]
- 4.Yao Y., Salas A., Logan I., Bandelt H.-J. mtDNA Data Mining in GenBank Needs Surveying. Am. J. Hum. Genet. 2009;85:922–933. doi: 10.1016/j.ajhg.2009.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vianello D., Sevini F., Castellani G., Laura L., Capri M., Franceschi C. HAPLOFIND: A new method for high-throughput mtDNA haplogroup assignment. Hum. Mutat. 2013;34:1189–1194. doi: 10.1002/humu.22356. [DOI] [PubMed] [Google Scholar]
- 6.Navarro-Gomez D., Leipzig J., Shen L., Lott M., Stassen A.P.M., Wallace D.C., Wiggs J.L., Falk M.J., van Oven M., Gai X. Phy-Mer: a novel alignment-free and reference-independent mitochondrial haplogroup classifier. Bioinformatics. 2015;31:1310–1312. doi: 10.1093/bioinformatics/btu825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fan L., Yao Y.-G. MitoTool: a web server for the analysis and retrieval of human mitochondrial DNA sequence variations. Mitochondrion. 2011;11:351–356. doi: 10.1016/j.mito.2010.09.013. [DOI] [PubMed] [Google Scholar]
- 8.Röck A.W., Dür A., van Oven M., Parson W. Concept for estimating mitochondrial DNA haplogroups using a maximum likelihood approach (EMMA) Forensic Sci. Int. Genet. 2013;7:601–609. doi: 10.1016/j.fsigen.2013.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Behar D.M., van Oven M., Rosset S., Metspalu M., Loogväli E.-L., Silva N.M., Kivisild T., Torroni A., Villems R. A ‘Copernican’ reassessment of the human mitochondrial DNA tree from its root. Am. J. Hum. Genet. 2012;90:675–684. doi: 10.1016/j.ajhg.2012.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bandelt H.-J., Kloss-Brandstätter A., Richards M.B., Yao Y.-G., Logan I. The case for the continuing use of the revised Cambridge Reference Sequence (rCRS) and the standardization of notation in human mitochondrial DNA studies. J. Hum. Genet. 2013;59:66–77. doi: 10.1038/jhg.2013.120. [DOI] [PubMed] [Google Scholar]
- 11.Skonieczna K., Malyarchuk B., Jawień A., Marszałek A., Banaszkiewicz Z., Jarmocik P., Borcz M., Bała P., Grzybowski T. Heteroplasmic substitutions in the entire mitochondrial genomes of human colon cells detected by ultra-deep 454 sequencing. Forensic Sci. Int. Genet. 2015;15:16–20. doi: 10.1016/j.fsigen.2014.10.021. [DOI] [PubMed] [Google Scholar]
- 12.Bandelt H.-J., Salas A. Current Next Generation Sequencing technology may not meet forensic standards. Forensic Sci. Int. Genet. 2012;6:143–145. doi: 10.1016/j.fsigen.2011.04.004. [DOI] [PubMed] [Google Scholar]
- 13.Just R.S., Irwin J.A., Parson W. Mitochondrial DNA heteroplasmy in the emerging field of massively parallel sequencing. Forensic Sci. Int. Genet. 2015;18:131–139. doi: 10.1016/j.fsigen.2015.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kloss-Brandstätter A., Weissensteiner H., Erhart G., Schäfer G., Forer L., Schönherr S., Pacher D., Seifarth C., Stöckl A., Fendt L., et al. Validation of next-generation sequencing of entire mitochondrial genomes and the diversity of mitochondrial DNA mutations in oral squamous cell carcinoma. PLoS One. 2015;10:e0135643. doi: 10.1371/journal.pone.0135643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.The 1000 Genomes Project Consortium. A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.van Oven M. PhyloTree Build 17: growing the human mitochondrial DNA tree. Forensic Sci. Int. Genet. Suppl. Ser. 2015;5:392–394. [Google Scholar]
- 17.King J.L., Sajantila A., Budowle B. mitoSAVE: mitochondrial sequence analysis of variants in Excel. Forensic Sci. Int. Genet. 2014;12:122–125. doi: 10.1016/j.fsigen.2014.05.013. [DOI] [PubMed] [Google Scholar]
- 18.Danecek P., Auton A., Abecasis G., Albers C.A., Banks E., DePristo M.A., Handsaker R.E., Lunter G., Marth G.T., Sherry S.T., et al. The variant call format and VCFtools. Bioinformatics. 2011;27:2156–2158. doi: 10.1093/bioinformatics/btr330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li S., Besenbacher S., Li Y., Kristiansen K., Grarup N., Albrechtsen A., Sparsø T., Korneliussen T., Hansen T., Wang J., et al. Variation and association to diabetes in 2000 full mtDNA sequences mined from an exome study in a Danish population. Eur. J. Hum. Genet. 2014;22:1040–1045. doi: 10.1038/ejhg.2013.282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ding J., Sidore C., Butler T.J., Wing M.K., Qian Y., Meirelles O., Busonero F., Tsoi L.C., Maschio A., Angius A., et al. Assessing mitochondrial DNA variation and copy number in lymphocytes of ∼2,000 sardinians using tailored sequencing analysis tools. PLOS Genet. 2015;11:e1005306. doi: 10.1371/journal.pgen.1005306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Excoffier L., Lischer H.E.L. Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resour. 2010;10:564–567. doi: 10.1111/j.1755-0998.2010.02847.x. [DOI] [PubMed] [Google Scholar]
- 22.Swafford D.L. PAUP*: Phylogenetic Analysis Using Parsimony(*and Other Methods) 2003. Version 4. Sinauer Associates, Sunderland, Massachusetts.
- 23.Huelsenbeck J.P., Ronquist F. MRBAYES: bayesian inference of phylogenetic trees. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- 24.Deza E., Deza M.M. Encyclopedia of Distances. Berlin; Heidelberg: Springer; 2009. [Google Scholar]
- 25.Bandelt H.-J., Salas A, Lutz-Bonengel S. Artificial recombination in forensic mtDNA population databases. Int. J. Legal Med. 2004;118:267–273. doi: 10.1007/s00414-004-0455-2. [DOI] [PubMed] [Google Scholar]
- 26.Brandstätter A., Sänger T., Lutz-Bonengel S., Parson W., Béraud-Colomb E., Wen B., Kong Q.-P., Bravi C.M., Bandelt H.-J. Phantom mutation hotspots in human mitochondrial DNA. Electrophoresis. 2005;26:3414–3429. doi: 10.1002/elps.200500307. [DOI] [PubMed] [Google Scholar]
- 27.Kong Q.-P., Salas A., Sun C., Fuku N., Tanaka M., Zhong L., Wang C.-Y., Yao Y.-G., Bandelt H.-J. Distilling artificial recombinants from large sets of complete mtDNA genomes. PLoS One. 2008;3:e3016. doi: 10.1371/journal.pone.0003016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Weissensteiner H., Forer L., Fuchsberger C., Schöpf B., Kloss-Brandstätter A., Specht G., Kronenberg F., Schönherr S. mtDNA-Server: next-generation sequencing data analysis of human mitochondrial DNA in the cloud. Nucleic Acids Res. 2016 doi: 10.1093/nar/gkw247. doi:10.1093/nar/gkw247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brandon M.C., Ruiz-Pesini E., Mishmar D., Procaccio V., Lott M.T., Nguyen K.C., Spolim S., Patil U., Baldi P., Wallace D.C. MITOMASTER: a bioinformatics tool for the analysis of mitochondrial DNA sequences. Hum. Mutat. 2009;30:1–6. doi: 10.1002/humu.20801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lott M.T., Leipzig J.N., Derbeneva O., Xie H.M., Chalkia D., Sarmady M., Procaccio V., Wallace D.C. mtDNA Variation and Analysis Using Mitomap and Mitomaster. Curr Protoc Bioinformatics. 2013;44:1.23.1–1.23.26. doi: 10.1002/0471250953.bi0123s44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Andrews R.M., Kubacka I., Chinnery P.F., Lightowlers R.N., Turnbull D.M., Howell N. Reanalysis and revision of the Cambridge reference sequence for human mitochondrial DNA. Nat. Genet. 1999;23:147. doi: 10.1038/13779. [DOI] [PubMed] [Google Scholar]
- 32.Ilyas M., Kim J.-S., Cooper J., Shin Y.-A., Kim H.-M., Cho Y.S., Hwang S., Kim H., Moon J., Chung O., et al. Whole genome sequencing of an ethnic Pathan (Pakhtun) from the north-west of Pakistan. BMC Genomics. 2015;16:1–8. doi: 10.1186/s12864-015-1290-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bandelt H.-J., van Oven M., Salas A. Haplogrouping mitochondrial DNA sequences in Legal Medicine/Forensic Genetics. Int. J. Legal Med. 2012;126:901–916. doi: 10.1007/s00414-012-0762-y. [DOI] [PubMed] [Google Scholar]
- 34.Bandelt H.-J., Quintana-Murci L., Salas A., Macaulay V. The fingerprint of phantom mutations in mitochondrial DNA data. Am. J. Hum. Genet. 2002;71:1150–1160. doi: 10.1086/344397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Soares P., Ermini L., Thomson N., Mormina M., Rito T., Röhl A., Salas A., Oppenheimer S., Macaulay V., Richards M.B. Correcting for purifying selection: an improved human mitochondrial molecular clock. Am. J. Hum. Genet. 2009;84:740–759. doi: 10.1016/j.ajhg.2009.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pereira L., Soares P., Radivojac P., Li B., Samuels D.C. Comparing phylogeny and the predicted pathogenicity of protein variations reveals equal purifying selection across the global human mtDNA diversity. Am. J. Hum. Genet. 2011;88:433–439. doi: 10.1016/j.ajhg.2011.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gómez J., García L.J., Salazar G. a., Villaveces J., Gore S., García A., Martín M.J., Launay G., Alcántara R., Del-Toro N., et al. BioJS: an open source JavaScript framework for biological data visualization. Bioinformatics. 2013;29:1103–1104. doi: 10.1093/bioinformatics/btt100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lischer H.E.L., Excoffier L. PGDSpider: an automated data conversion tool for connecting population genetics and genomics programs. Bioinformatics. 2012;28:298–299. doi: 10.1093/bioinformatics/btr642. [DOI] [PubMed] [Google Scholar]
- 39.Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Beerli P., Palczewski M. Unified framework to evaluate panmixia and migration direction among multiple sampling locations. Genetics. 2010;185:313–326. doi: 10.1534/genetics.109.112532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Felsenstein J. PHYLIP - Phylogeny Inference Package (Version 3.2) Cladistics. 1989;5:164–166. [Google Scholar]
- 42.Maddison D.R., Swofford D.L., Maddison W.P. NEXUS: an extensible file format for systematic information. Syst. Biol. 2009;46:590–621. doi: 10.1093/sysbio/46.4.590. [DOI] [PubMed] [Google Scholar]
- 43.Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M.A.R., Bender D., Maller J., Sklar P., de Bakker P.I.W., Daly M.J., et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.