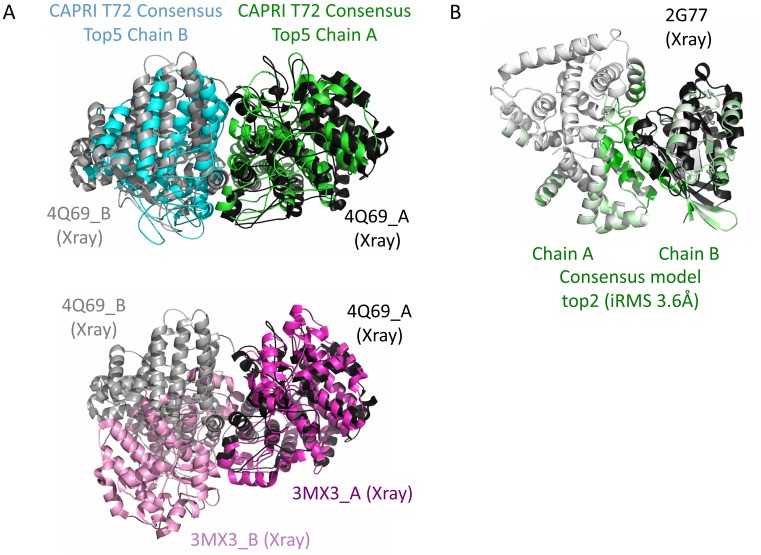
Figure 2.
Successful examples from a CAPRI challenge and from Weng benchmark v4. (A) The challenge for CAPRI target T72 (round 30) was to predict the homodimeric assembly of a protein whose monomeric structure could be modeled at 20% sequence identity using the template 3MX3 (pink structure). Modeling the homodimer using template-based approach based on 3MX3 dimeric assembly (pink) would have led to incorrect prediction since the experimental structure of T72 (4Q69 shown in black and gray) was eventually found to assemble in a different manner. Still, during CAPRI round 30 our group obtained the lowest iRMSD of 3.5Å for this prediction which was only well predicted by two other methods (Haddock and SwarmDock). InterEvDock server successfully identified in its top10 consensus an acceptable model rank as top5 (cyan and green model). (B) 2G77 example from Weng benchmark v4 (21). The model is shown as white and green cartoon while the X-ray structure is in black with only the B chain displayed. Green color reports for the likelihood of a residue to be at the interface calculated from the consensus method over the 3xtop10 decoys. It supports the prediction of the 10 most likely residues at interface provided by the server which is very precise for 2G77 with all 10 predicted residues at interface. InterEvDock consensus detected an acceptable model ranked top2 although it was missed by SwarmDock. This figure illustrates that the residues predicted to lie in the consensus interface provide very good hints to guide the mutagenesis.