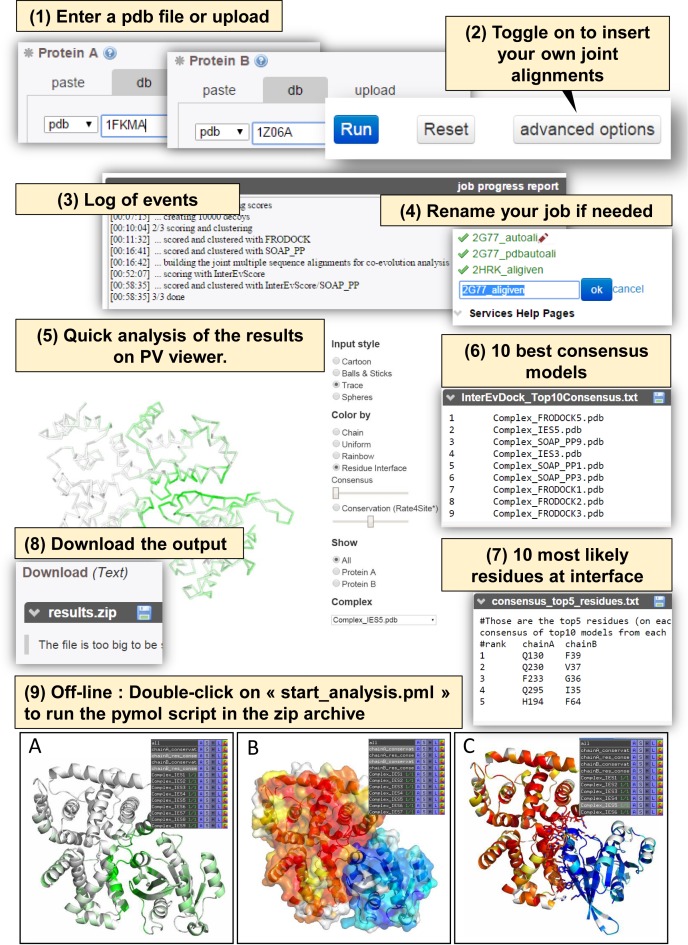
Figure 3.
Example of input and output in the InterEvDock server. InterEvDock integrates a number of important features that are indexed from 1 to 9. (1) Input pdbs can be either uploaded or retrieved automatically by specifying the pdb code and the chain. (2) Multiple sequence alignments can be either automatically generated on the server or uploaded by clicking on this option. (3) A log allows following the time required for the different steps of the docking. (4) Jobs can be easily renamed in the left panel at any time. (5) At the end of the docking process, the top10 decoys for every scoring methods can be analyzed in the PV viewer. Several coloring options are proposed to map either the probability of a residue to be at interface (from green to white) or the conservation index as calculated by rate4site (32,33) (from red to white through yellow). (6) List of the 10 best consensus models predicted by InterEvDock server. (7) List of the 10 most likely residues at interface predicted from the consensus analysis. (8) Zip archive containing all the top50 models selected by InterEvScore, SOAP-PP and FRODOCK. (9) A ‘start_analysis.pml’ pymol script is distributed in every zip archive which can be opened in a single click to generate pre-processed views of the 3xtop10 models, including color mapping schemes as in (5).