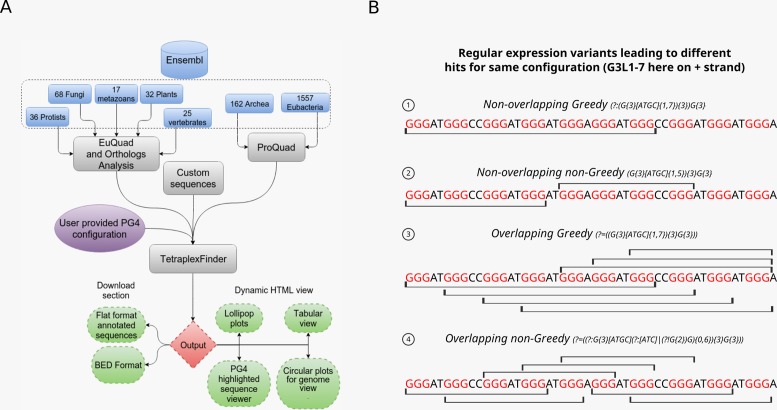
Figure 1.
Work flow and algorithms of PG4 motif mining (A) Schematic diagram depicting the workflow of QuadBase2 web server. Users interact with either EuQuad or ProQuad interface to select organisms and genes of interest. Information on genes and their orthologues is obtained from the current release of Ensembl. Here, the number of species from each meta-class in the current release of QuadBase2 are shown. TetraplexFinder works both as an interface and the core algorithm that performs PG4 motif mining on the server backend. The PG4 motif configurations are provided by the user in all three modules through an interactive interface. The result of PG4 motif mining can either be downloaded or visualized interactively for each gene. (B) Figure depicting the results of PG4 algorithms on an example sequence. The line over and below each sequence shows the PG4 motif that would be captured by the algorithm. The ‘greedy’ and ‘non-greedy’ algorithms detect different number and lengths of PG4 motifs. Choice of algorithm is crucial for downstream interpretation.