Fig. 1.
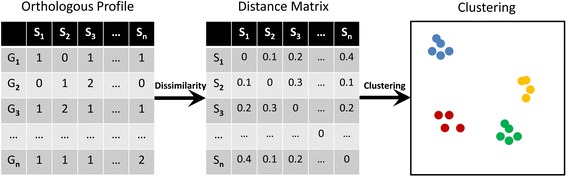
The flowchart of applying gene content dissimilarity for microbial delineation and classification. Three main steps were included. First, orthologous gene profiles were obtained for all selected microbial genomes by searching against the eggNOG database. Second, pairwise gene content dissimilarity as measured by Bray-Curtis dissimilarity was calculated for all pairs of microbial strains. Third, microbial strains were clustered into different groups
