ABSTRACT
Paramyxoviridae consist of a large family of enveloped, negative-sense, nonsegmented single-stranded RNA viruses that account for a significant number of human and animal diseases. The fusion process for nearly all paramyxoviruses involves the mixing of the host cell plasma membrane and the virus envelope in a pH-independent fashion. Fusion is orchestrated via the concerted action of two surface glycoproteins: an attachment protein called hemagglutinin-neuraminidase (HN [also called H or G depending on virus type and substrate]), which acts as a receptor binding protein, and a fusion (F) protein, which undergoes a major irreversible refolding process to merge the two membranes. Recent biochemical evidence suggests that receptor binding by HN is dispensable for cell-cell fusion. However, factors that influence the stability and/or conformation of the HN 4-helix bundle (4HB) stalk have not been studied. Here, we used oxidative cross-linking as well as functional assays to investigate the role of the structurally unresolved membrane-proximal stalk region (MPSR) (residues 37 to 58) of HN in the context of headless and full-length HN membrane fusion promotion. Our data suggest that the receptor binding head serves to stabilize the stalk to regulate fusion. Moreover, we found that the MPSR of HN modulates receptor binding and neuraminidase activity without a corresponding regulation of F triggering.
IMPORTANCE Paramyxoviruses require two viral membrane glycoproteins, the attachment protein variously called HN, H, or G and the fusion protein (F), to couple host receptor recognition to virus-cell fusion. The HN protein has a globular head that is attached to a membrane-anchored flexible stalk of ∼80 residues and has three activities: receptor binding, neuraminidase, and fusion activation. In this report, we have identified the functional significance of the membrane-proximal stalk region (MPSR) (HN, residues 37 to 56) of the paramyxovirus parainfluenza virus (PIV5), a region of the HN stalk that has not had its structure determined by X-ray crystallography. Our data suggest that the MPSR influences receptor binding and neuraminidase activity via an indirect mechanism. Moreover, the receptor binding head group stabilizes the 4HB stalk as part of the general mechanism to fine-tune F-activation.
INTRODUCTION
Paramyxoviruses are enveloped, nonsegmented negative-stranded RNA viruses that cause diseases in humans and animals. Notable members of this family of viruses include parainfluenza viruses 1 to 5 (PIV1 to -5), Newcastle disease virus (NDV), mumps virus, measles virus, Nipah virus, Hendra virus, respiratory syncytial virus (RSV), and human metapneumovirus. For infection of cells to occur, the virus and host cell membranes fuse at neutral pH (1–13) to deliver the viral ribonucleoprotein into the cytoplasm. The fusion of nearly all paramyxoviruses requires the concerted action of two viral surface glycoproteins: the hemagglutinin-neuraminidase (HN [also called H or G depending on virus type]) and a fusion protein (F). The involvement of the HN protein in receptor binding and neuraminidase (NA) activity as well as fusion promotion has been well documented (14–18). However, the mechanistic details by which HN causes F activation remain poorly defined. PIV5 HN binds to cell surface sialic acid, which brings the cell and virus membrane in close proximity. It is thought that HN binding to its ligand triggers the activation of the fusion protein, which is kinetically trapped in a metastable state (19–21), to undergo a large-scale irreversible refolding event that culminates in virus-cell membrane merger (22–25).
HN is a type II membrane protein with an N-terminal stalk (residues 1 to 117) and a globular head that contains the NA active site (residues 118 to 565) (26–28). The ectodomain stalk comprises residues 37 to 118, and HN is anchored into the viral membrane by the transmembrane domain. The cytoplasmic tail of HN has been shown to play a role in fusion promotion (29–31), but the mechanism of action is unknown.
The atomic structure of the stalk revealed a 4-helix bundle (4HB) with an upper portion of the stalk implicated as the F-activating region (FAR) that mediates activation (16, 17). The “provocateur” model of paramyxovirus fusion provides a compelling explanation of how sialic acid binding by the head group of HN transmits the signal for F activation (15, 32). This model suggests that in the absence of receptor binding, the FAR at the membrane-distal end of the HN is inaccessible for F interaction. However, upon receptor engagement the bulky head groups move to the “up” position, relieving the steric obstruction and facilitating a physical interaction between HN and F (18, 20, 33). It has been shown that the HN1-117 alone (headless stalk) promotes F activation to levels commensurate with the wild-type (WT) HN and even higher levels at elevated temperatures (2, 17, 20). Recently, the Ig-like domain of the trimeric F protein been implicated in the F-HN interaction (2).
For many paramyxoviruses, an interaction between F and HN, H, or G has been shown by either coimmunoprecipitation or intracellular coretention of the two proteins (34–41). However, this has proven difficult to demonstrate for PIV5 F and HN, and it has been postulated that an F-HN interaction is either transient or F and HN associate weakly. Although the “headless HN stalk” promotes F activation, it has been found that the length of the HN1-117 stalk affects the level of fusion (33, 42, 43). The mechanistic explanation for this phenomenon is not known, but presumably, alterations in the stalk length affect its conformation or orientation or the dynamics of the FAR in a way that disfavors fusion with its cognate F protein.
The atomic structure of PIV5 HN1-117 (residues 56 to 117) reveals a 4-helix bundle (4HB) structure in which the membrane-distal section of the stalk adopts an 11-mer repeat with a less coiled conformation and the membrane-proximal part of the stalk adopts a left-handed superhelical twist. The structure of the adjacent membrane-proximal stalk region (MPSR) (residues 37 to 58) remains unsolved but has been predicted to be α-helical (44). This region may play an important regulatory role such as in oligomerization and stability of the stalk and hence govern the timely interaction of F and HN. The amino acid at the purported boundary of the PIV5 TM and ectodomain (E37) was found to regulate the rate of internalization of HN (45). However, the role of the primary sequence in fusion promotion, receptor binding, and NA activity has not been explored. Unlike the TM region of HN, which is rich in hydrophobic residues, the adjoining MPSR is mainly rich in charged residues that may be involved in stabilizing interactions such as hydrogen bonds or salt bridges. Also, this region may function as a molecular recognition system—for example, to regulate protein internalization and general turnover (45).
In this report, we investigated the structure-function role of PIV5 HN MPSR in receptor binding, NA activity, and fusion promotion. By using cysteine mutagenesis and oxidative cross-linking of PIV5 HN, we show that the MPSR arrangement differs in the headless HN from the full-length HN protein. The receptor binding head stabilizes the stalk and regulates F activation. In addition, we found that residues in the MPSR can modulate receptor binding and NA activity without changing the fusion promotion properties of HN.
MATERIALS AND METHODS
Cells and antibodies.
HEK 293T, and Vero cells were grown and maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and 1% penicillin–streptomycin. BHK-21F cells were grown in DMEM containing 10% FBS and 10% tryptose phosphate broth. For BSR-T7/5 cells (a BHK clone expressing T7 RNA polymerase), Geneticin G418 (500 μg/ml) was added during every third passage in DMEM–10% FBS. Immunoprecipitation of PIV5-HN full-length and HN1-117 stalk was done using a combination of the monoclonal antibodies (MAb) HN1b and HN4b or polyclonal antibody (pAb) R471 as described previously (20).
Cloning and mutagenesis.
pCAGGS-HN and pCAGGS-F expression constructs harboring the PIV5 (W3A) F and HN genes were used as described previously (20). The PIV5 HN1-117 construct (HN1-117) was created by PCR amplification of the stalk region (residues 1 to 117) from the full-length PIV5 HN (residues 1 to 565). The PCR fragment was then cloned into the pmEGFP-C1 vector using SacI and KpnI sites. Single and triple (3×) alanine mutations as well as cysteine mutations were constructed using the QuikChange mutagenesis kit (Agilent Technologies, Santa Clara, CA) according to the manufacturer's instructions. The resultant mutants were subcloned into pCAGGS vector for mammalian cell expression. The nucleotide sequence of the entire open reading frame for each mutant was verified using an Applied Biosystems 3100-Avant automated DNA sequencer (Life Technologies Corp., Carlsbad, CA).
Immunoprecipitation and SDS-PAGE.
To examine expression of HN WT, HN1-117, and mutant proteins, 293T cells were transfected with pCAGGS-HN and HN mutant plasmids using a standard transfection protocol. After 16 posttransfection (p.t.), proteins were metabolically labeled with 35S label and immunoprecipitated as described previously (16). Polypeptides were analyzed under reducing and nonreducing conditions using 10% or 17.5% SDS-PAGE gel depending on the protein mass. To determine the tetramer/dimer ratio of HN, 293T cells expressing WT HN, HN1-117, and HN cysteine mutants were treated with copper(II) phenanthroline (CuP) as described previously (46). Briefly, after 35S labeling, cells were subjected to three freeze-thaw cycles followed by CuP labeling before lysis and immunoprecipitation with MAb HN1b and HN4b for the full-length HN and pAb R471 for headless HN.
Flow cytometry.
The cell surface expression of the HN WT and HN mutants was assessed using flow cytometry as described previously (16). Briefly, overnight-transfected cells were washed and incubated with a 1:100 dilution of HN pAb R471 or MAb HN4b at 4°C for 1 h followed by incubation with 1:100 dilution of fluorescein isothiocyanate (FITC)-conjugated goat anti-rabbit IgG or goat anti-mouse IgG (Jackson ImmunoResearch, West Grove, PA) as the secondary antibody. Fluorescence intensity was quantified using a FACSCaliber flow cytometer (Becton Dickinson, Franklin Lakes, NJ).
HAd assay.
To measure the receptor binding activity of both WT HN and HN mutants, monolayer cultures of 293T cells were transfected with pCAGGS-HN and HN mutants (1 μg plasmid DNA each). At 18 h p.t., cells were washed gently with ice-cold PBS+ (phosphate-buffered saline containing calcium and magnesium) followed by incubation for 2 h with 1% chicken erythrocytes (RBCs) in PBS+ at 4°C to allow for receptor binding (but not fusion). Cells were washed with cold PBS+ five times to remove unbound RBCs. Bound RBCs were lysed in 0.5 ml ice-cold distilled water and rocked for 2 h at 4°C. The lysate was cleared by centrifugation (5,000 rpm for 2 min), and the absorbance of the supernatant was read at 410 nm using a Beckman Coulter DU 730 Life Sciences UV-visible (UV-Vis) spectrophotometer (Beckman Coulter, Brea, CA). The hemadsorption (HAd) assay was also repeated under reducing conditions. Prior to RBC incubation, cells were treated for 30 min at 37°C with 10 mM tris(2-carboxyethylene) phosphine (TCEP) in PBS.
NA activity assay.
293T cells were transfected with WT HN and HN mutants (1 μg plasmid DNA each) using the Lipofectamine LTX reagent (Thermo Fisher Scientific) according to the manufacturer's protocol. Cells were detached from the plates using 500 μl/well 530 μM EDTA in PBS. The cells were centrifuged at 5,000 rpm for 3 min. The pellets were washed and resuspended in PBS+ and pelleted for 3 min at 5,000 rpm at 4°C. The cell pellets were then resuspended in 100 μl of 125 mM sodium acetate buffer (pH 4.75) containing 6.25 mM CaCl2. Twenty-five microliters of 5 mM 4-methylumbelliferyl-N-acetyl-α-d-neuraminic acid (Sigma-Aldrich, St. Louis, MO) was added to the reaction mixture. The reaction was allowed to proceed for 30 min at 37°C with occasional mixing. Seventy-five microliters of 20 mM sodium carbonate buffer (pH 10.4) was added to stop the reaction. Cells were pelleted, and 180 μl of the supernatant was transferred to a 96-well plate. Fluorescence of the cleaved substrate was measured at excitation and emission wavelengths of 356 and 450 nm, respectively, using a Spectramax M5 plate reader (Molecular Devices, Sunnyvale, CA). The assay was also repeated under reducing conditions using 10 mM TCEP.
Syncytium formation.
BHK-21F cells were transfected with pCAGGS-F and pCAGGS-HN or HN mutant plasmids, including HN1-117 stalk mutants (1 μg each plasmid DNA). Cells were washed with PBS after 18 h p.t, fixed, and stained using a Hema3 staining protocol (Fisher Scientific, Pittsburgh, PA) according to the manufacturer's instructions. The monolayers were photographed using an inverted phase-contrast microscope (Diaphot; Nikon, Melville, NY) connected to a digital camera (DCS 760; Kodak, Rochester, NY).
Luciferase reporter fusion assay.
To quantify the fusion of WT HN and HN1-117 mutants, monolayers of Vero cells (70 to 80% confluence) were transfected with 1 μg each pCAGGS-F, pCAGGS-HN, and pT7-luciferase, a plasmid that expresses firefly luciferase under T7 polymerase control. BSR-T7/5 cells, expressing T7 RNA polymerase, were removed with 50 mM EDTA and overlaid on the Vero cell monolayer at 16 h following transfection and incubated further for 6 h at 37°C. Unbound BSR-T7/5 cells were washed with PBS. Reporter Glo lysis buffer (Promega, Madison WI) was used to lyse the cells. Subsequently, the cell lysates were pelleted by centrifugation (10,000 rpm for 1 min), and 150 μl of the cleared lysates was then added to a 96-well dish along with 150 μl of the luciferase assay substrate (Promega). The luciferase activities of both the WT and HN mutants were quantified using a SpectraMax M5 plate reader (Molecular Devices). Luciferase activity was expressed as relative light units (RLU).
RESULTS
Role of MPSR residues in fusion promotion of the headless HN stalk.
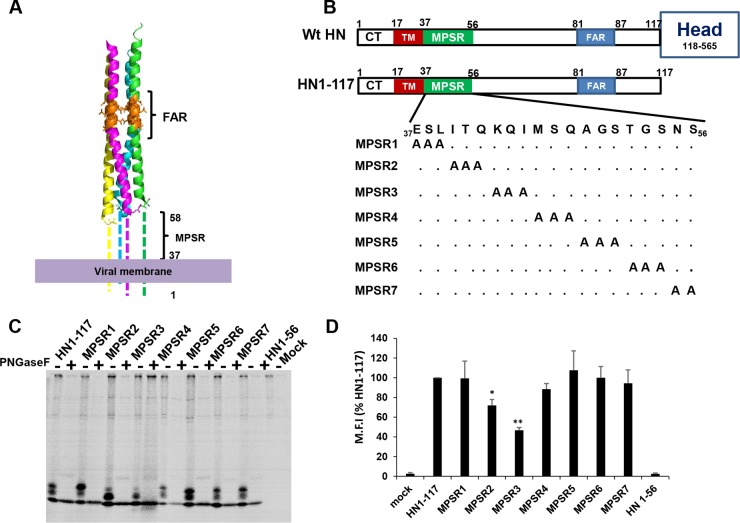
The structure and function of the MPSR of paramyxovirus HN/H/G are largely unknown. The atomic structures of paramyxovirus HNs do not reveal data for the MPSR (16, 18). To investigate the function of the MPSR of PIV5 HN, Ala scanning mutagenesis was used to generate triple (3×) Ala mutants for the first 20 HN residues (residues 37 to 56) of the MPSR of the headless HN stalk, HN1-117 (Fig. 1A and B). Earlier studies have shown HN1-117 to promote fusion of F (17, 20). Each HN1-117 3× Ala MPSR mutant was tested for its expression and fusion promotion. Protein expression of the HN1-117 3× Ala mutants was evaluated using 35S metabolic labeling followed by immunoprecipitation as described in Materials and Methods. When the immune-precipitated proteins were treated with peptide-N-glycosidase F (PNGase F) to remove carbohydrate chains, all of the mutants were susceptible to digestion to a single species except HN1-56, which was either not expressed or more likely not recognized by the antibody (Fig. 1C). HN1-117 is predicted to contain a single site for addition of N-linked glycosylation at residue 110, and therefore the three gel bands most likely represent two forms of glycosylation and unglycosylated protein. Flow cytometry was used to quantify the levels of cell surface expression of the HN1-117 3× Ala mutants. As shown in Fig. 1D, all the HN1-117 3× Ala mutants were expressed at the cell surface except HN1-56. It was noted that whereas mutants MPSR2 and MPSR3 were reduced in surface expression compared to HN1-117, all the other mutants were expressed at the cell surface in nearly similar amounts.
FIG 1.
Expression of MPSR triple alanine mutants. The 3× Ala mutants were made by mutations at the first 20 residues of the ectodomain of PIV5 headless HN1-117 to determine its functional significance. (A) Atomic model of PIV5 headless HN1-117 showing unresolved membrane-proximal stalk region (MPSR). F-interacting region (FAR) at the upper portion of the HN stalk is indicated. (B) Schematic diagram illustrating the PIV5 HN protein and showing the different domains: F-interacting region (blue), membrane-proximal stalk region (green), and transmembrane (TM) domain (red). The scheme of the 3× Ala mutants is shown. (C) Protein expression was determined using 30 min of 35S labeling followed by immunoprecipitation. Glycosylated species were digested by peptide-N-glycosidase F ([PNGase F]) treatment. Polypeptides were analyzed by 17.5% SDS-PAGE under reducing conditions. (D) Cell surface expression of HN1-117 and 3× Ala mutants was determined by flow cytometry. M.F.I., mean fluorescent intensity. Values are expressed as a percentage of HN1-117. Error bars represent standard deviations from three experiments. P values were calculated using Student's t test. *, P < 0.05; **, P < 0.01; otherwise, P > 0.05.
Fusion promotion by HN1-117 3× Ala MPSR mutants.
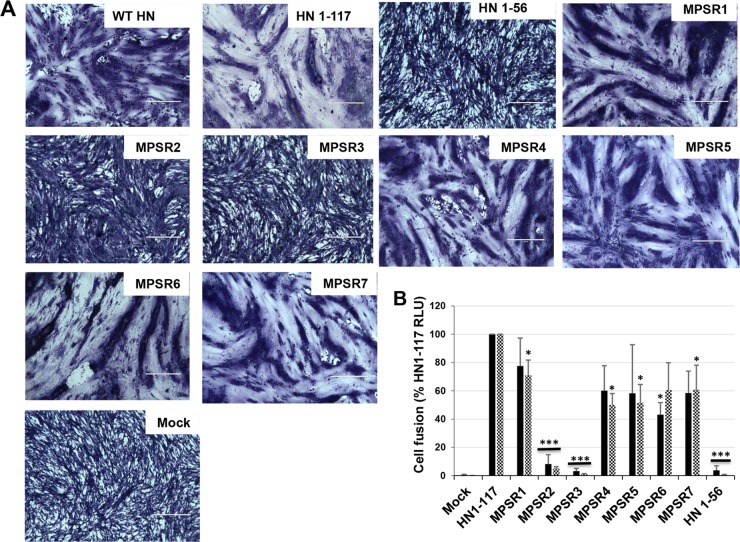
It has been shown previously that the HN headless stalk of PIV5 HN (HN1-117) and the stalks of other paramyxoviruses can activate their cognate F protein (17, 20, 43, 47, 48). To examine the effect of the HN1-117 3× Ala mutants on syncytium fusion, BHK cells were transfected with plasmids to express WT F and HN1-117 or the HN1-117 3× Ala mutants. As shown in Fig. 2A, all the HN1-117 3× mutants caused syncytium formation except MPSR2 and MPRS3.
FIG 2.
Fusogenic properties of HN1-117 MPSR 3× Ala mutants. (A) BHK cells were transfected with PIV5 F and HN1-117 or the respective triple mutants. Formation of syncytia was observed at 16 h p.t., and the cells were stained. (B) Quantitative luciferase activity (fusion) of both HN1-117 and 3× Ala mutants. Fusion was performed at 33°C (black bars) or 37°C (checkered bars). Luciferase activity was expressed as relative light units (RLU) of HN1-117. Error bars represent standard deviations from three independent experiments. Statistical significance of difference between each mutant and HN1-117 was assessed using Student's t test. *, P < 0.05; ***, P < 0.001; otherwise, P > 0.05.
To quantify the fusion activity of the HN1-117 3× Ala mutants, a luciferase fusion assay (16) was used. Vero cells were transiently transfected with HN1-117 3× Ala MPSR mutants together with pCAGGS F and pT-7 luciferase after overnight-transfected cells were washed with PBS+ and overlaid with BSR-7 cells. As shown in Fig. 2B, all the mutants promoted cell-cell fusion at both 33 and 37°C except MPSR 2 and MPSR3, where fusion promotion activity was negligible. Taken together, the data suggest that residues at positions 40 to 45 of HN1-117 may play a role in fusion promotion by the HN stalk.
Effect of single point HN1-117 MPSR mutations on fusion.
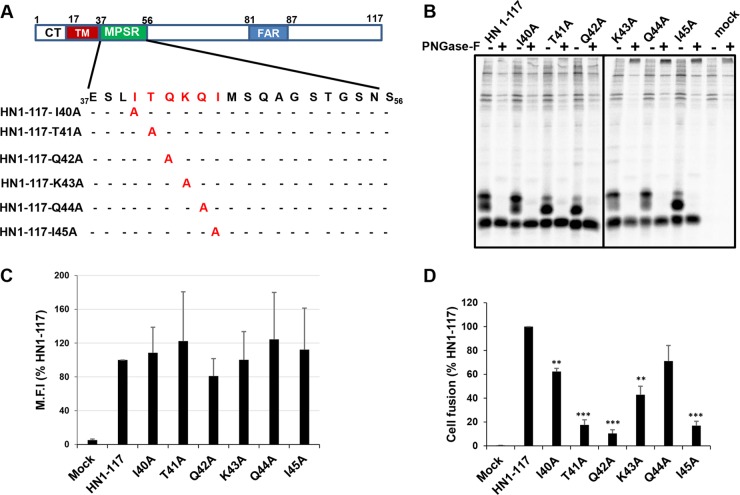
To determine the residues in the headless HN1-117 responsible for the absence of fusion promotion, individual residues at residues 40 to 45 (comprising MPSR2 and MPSR3) were substituted for with Ala and characterized (Fig. 3A). All the HN1-117 Ala point mutants were expressed and on PNGase F digestion, all the mutants yielded a single species (Fig. 3B). Differences in the amounts of the two species modified by carbohydrate chains were observed as well as subtle shifts in electrophoretic mobility that may reflect the amino acid substitutions.
FIG 3.
Effect of single point mutations of HN1-117 MPSR on fusion promotion. (A) Schematic diagram of HN1-117 showing positions of single Ala point mutants. (B) Protein expression was analyzed by 35S metabolic labeling and radioimmunoprecipitation. Proteins were treated with or without PNGase F to assess susceptibility to digestion. Polypeptides were analyzed by 17.5% SDS-PAGE. (C) Cell surface expression of HN1-117 and single point Ala mutants was quantified by flow cytometry. (D) Fusion promotion of HN1-117 mutants were assessed using the luciferase fusion assay. Error bars represent standard deviations from three experiments. P values were calculated using paired Student's t test to determine statistical significance of difference between each mutant and HN1-117. **, P < 0.01; ***, P < 0.001; otherwise, P > 0.05.
Cell surface expression was analyzed by flow cytometry (Fig. 3C), and the data showed that all the point mutants were expressed at the cell surface similar to HN1-117. In contrast to cell surface expression levels, the fusion promotion activities, as measured by luciferase assay, of the HN1-117 T41A, Q42A, and I45A mutants were significantly reduced (Fig. 3D). The above observations suggest that residues T41, Q42, and I45 in HN1-117 play a role in fusion promotion. It is also noted that these three mutants do not accumulate the species with the slowest-migrating carbohydrate chain modification to the same level as HN1-117. These data suggest that the changes in the carbohydrate modification may influence the conformation and fusion promotion function of HN1-117.
Mutations in full-length HN MPSR affect receptor binding and NA but not fusion.
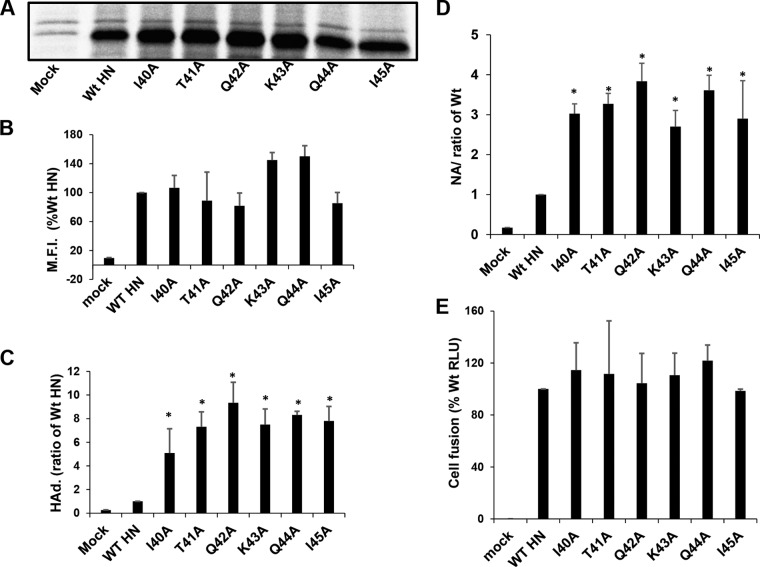
Given that mutations in the MPSR of the headless HN1-117 construct resulted in fusion defects, the same mutations were introduced into the full-length HN protein, and their effects on receptor binding, NA activity, and fusion promotion were evaluated. 293T cells were transfected with pCAGGS expressing full-length WT HN (residues 1 to 565) as well the HN proteins of the MPSR point Ala mutants. Protein expression was examined by 35S metabolic labeling and immunoprecipitation (Materials and Methods). It was observed that mutant HN proteins were expressed and transported to the cell surface at levels similar to that of the WT HN (Fig. 4A and B). Measurement of the HAd activity (Fig. 4C) and NA activity (Fig. 4D) indicated an ∼5- to 9-fold increase in each activity over WT HN. Unexpectedly, all the MPSR point mutants in full-length HN had comparable fusion promotion activity, unlike the fusion activation data obtained with the same mutations in HN1-117 stalk.
FIG 4.
Point mutations in the MPSR of full-length HN affect receptor binding and NA activity but not fusion promotion. The point mutations shown in Fig. 3 were made in full-length WT HN to assess its biological function. (A) 293T cells were transfected with pCAGGS vector encoding full-length WT HN or the MPSR point mutants. HN was metabolically 35S labeled and immunoprecipitated, and polypeptides were analyzed by 10% SDS-PAGE. (B) Cell surface expression of WT HN and the point Ala mutants. (C and D) NA activity and HAd activity of both WT HN and HN point mutants, respectively. Error bars are standard deviations from three independent experiments. (E) Fusion promotion by WT HN and mutants was quantified using the luciferase fusion assay. Student's t test was used to determine statistical significance. *, P < 0.05; otherwise, P > 0.05.
HN MPSR can form reversible disulfide bonds with limited functional defects.
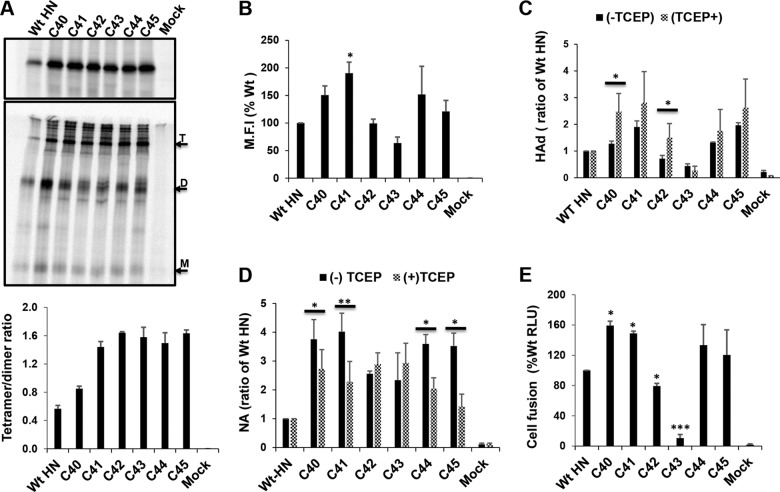
Full-length PIV5 HN is a dimer of dimers, with each dimer forming a disulfide bond at residue C111 (18). To probe if the HN MPSR residues are important for the conformational flexibility of the HN stalk, residues 40 to 45 were individually mutated to cysteine in full-length HN. 293T cells transiently expressing WT HN and the cysteine mutants were 35S metabolically labeled, and proteins were immunoprecipitated. Polypeptides were subjected to SDS-PAGE under reducing (Fig. 5A, top) and nonreducing (Fig. 5A, middle) conditions. The presence of a cysteine at residues 40 to 45 increased the HN tetramer/dimer ratio (Fig. 5A, bottom). The cell surface expression levels of the mutants were similar to those of WT HN except for C41, which showed a statistically significant higher level of surface expression than WT HN (Fig. 5B). Formation of disulfide-linked tetramers led to enhanced HAd (Fig. 5C) and NA (Fig. 5D) activity, depending on the position of the Cys residue. However, subsequent reduction with TCEP resulted in residue-specific increase in HAd (particularly, C40 and C42), while in most cases a reduction in NA was observed. Fusion promotion was not negatively impacted except for C43, which abolished fusion (Fig. 5E). The above observations suggest that the MPSR of PIV5 HN plays a role in conformational flexibility of the stalk and indirectly regulate receptor engagement.
FIG 5.
Disulfide-linked stabilization of HN MPSR. (A) 293T cells expressing HN with cysteine mutations at residues 40 to 45 were 35S metabolically labeled and HN immunoprecipitated, and polypeptides were analyzed by reducing (top) and nonreducing (middle) 10% SDS-PAGE. The different mobilities of WT HN and the mutants are indicated by the monomer (M), dimer (D), and tetramer (T). ImageJ was used to quantify band intensity, and the ratio of tetramer to dimer was estimated (bottom). (B) Cell surface expression of WT HN and cysteine mutants and (C) the effect of disulfide linked tetramerization of the MPSR on receptor binding (HAd) were measured under reducing (10 mM TCEP) and nonreducing conditions. Asterisks indicate statistical significance between TCEP treated and untreated. (D) NA activity of Cys mutants under reducing (TCEP) and nonreducing conditions. (E) Quantitative luciferase fusion assay of WT HN and HN cysteine mutants. *, P < 0.05; **, P < 0.01; ***, P < 0.001; otherwise, P > 0.05.
Formation of disulfide-linked headless and full-length HN.
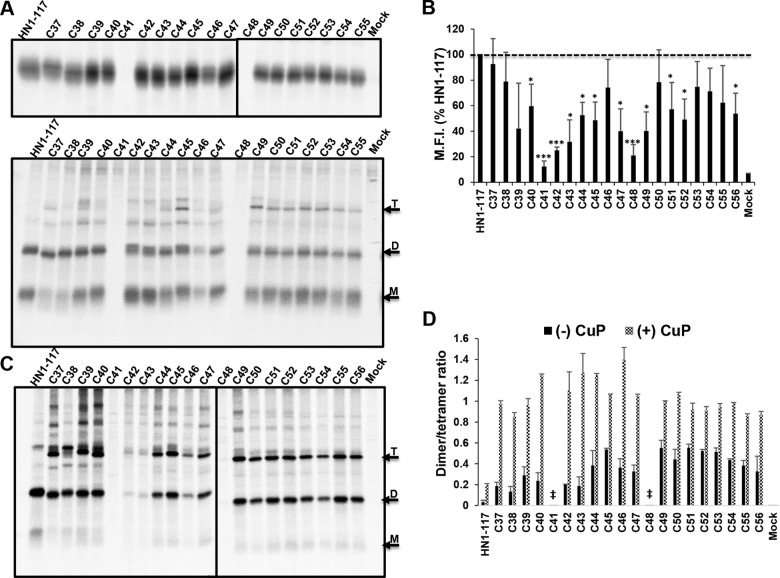
To investigate if disulfide bond formation would show a periodic pattern suggestive of a 4-helix bundle (1, 46, 49), the 20 residues of the MPSR (residues 37 to 56) were mutated to Cys in the HN1-117 stalk background. HEK 293T cells transiently expressing WT HN1-117 as well as the cysteine mutants were 35S metabolically labeled, the proteins were immunoprecipitated, and the polypeptides were subjected to electrophoresis on SDS-PAGE under reducing and nonreducing conditions. As shown in Fig. 6A (top and bottom), these residues showed limited propensity to form a tetramer under the nonreducing condition. A significant portion of the HN1-117 Cys mutants remained as monomers and disulfide-linked dimers and did readily form disulfide-linked tetramers. Although the vast majority of the Cys mutants were expressed at the cell surface to various degrees, two of these mutants (C41 and C48) were not detected when immunoprecipitated with HN polyclonal antibody. None of the Cys HN1-117 mutants was expressed at levels above HN1-117 (Fig. 6B). Interestingly, when the proteins were oxidized with CuP, a significant number of the HN1-117 Cys mutants (34–37, 39–44) showed increased tetramer formation (Fig. 6C). Protein bands were quantified with ImageJ software to determine the tetramer/dimer ratio shown in Fig. 6D.
FIG 6.
The stalk of HN1-117 forms limited disulfide-linked tetramers. MPSR residues 37 to 56 in HN1-117were substituted for with cysteine to determine if the stalk could be trapped via covalent disulfide bonds. (A) 293T cells expressing HN1-117 and Cys mutants were 35S labeled, HN was immunoprecipitated, and polypeptides were analyzed by 17.5% SDS-PAGE under reducing (top) and nonreducing (bottom) conditions. Arrows indicate HN1-117 monomer (M), dimer (D), and tetramer (T). (B) Cell surface expression was quantified by flow cytometry and expressed as mean fluorescent intensity (M.F.I.) of HN1-117. The horizontal dashed line represents the HN1-117 expression level. (C) 35S metabolically labeled 293T cells were treated with 3 μM (final concentration) copper(II) phenanthroline (CuP) prior to lysis and immunoprecipitation. Both HN1-117and Cys mutants were resolved by nonreducing 17.5% SDS-PAGE after PNGase F digestion to remove carbohydrate chains. (D) The ratio of tetramer to dimer was estimated by quantifying band intensities with ImageJ. Error bars are standard deviations from two experiments. ‡, tetramer/dimer ratios were not estimated for C41 and C48 due to lack of detectable signal on the gel. *, P < 0.05; ***, P < 0.001; otherwise, P > 0.05.
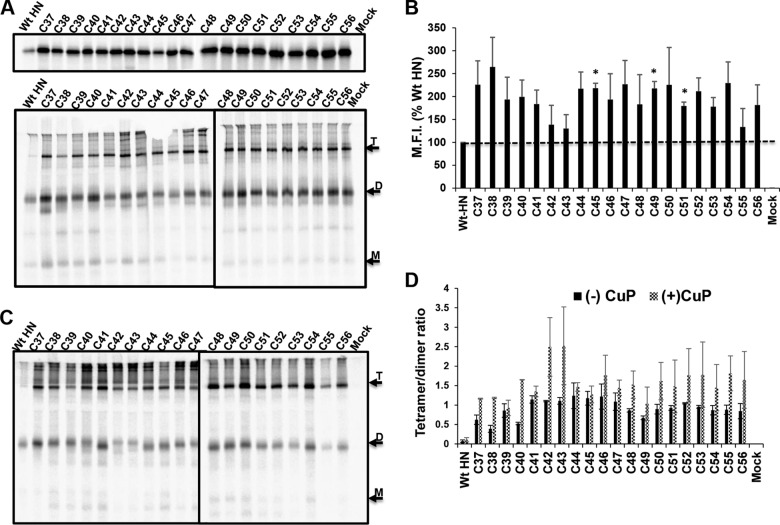
The introduction of cysteine mutations in the MPSR of full-length HN (Fig. 7A top, reducing; bottom, nonreducing) resulted in cell surface expression above WT HN levels (Fig. 7B). This finding with full-length HN is in contrast to the cell surface expression of the HN1-117 cysteine stalk mutants (Fig. 6A and B), where disulfide-linked stabilization did not improve cell surface expression. It can also be seen that the full-length HN protein readily forms disulfide-linked tetramers even in the absence of CuP oxidation (Fig. 7A, bottom, and C). To quantify the tetramer formation, protein band intensities were measured. As indicated in Fig. 7D, the tetramer/dimer ratio shows that the majority of the residues in the MPSR had a tetramer/dimer ratio of ≥1, but there was no detectable periodicity to the oxidation.
FIG 7.
The stalk of full-length HN can be trapped as disulfide-linked tetramers at the MPSR. (A) 35S metabolically labeled Cys mutants of full-length HN were resolved on a 10% SDS-PAGE under reducing (top) or nonreducing (bottom) conditions. Arrows indicate monomers (M), dimers (D), and tetramers (T) were 35S metabolically labeled. HN was immunoprecipitated, and polypeptides were analyzed by 10% SDS-PAGE. (B) Cell surface expression was quantified by flow cytometry and expressed as percentage of WT mean fluorescent intensity (M.F.I.). The horizontal dashed line represents the WT HN expression level. (C) WT HN and MPSR Cys mutants treated with CuP were resolved by nonreducing 10% SDS-PAGE. (D) Intensity bands of CuP-labeled proteins (C37 to C56) were quantified using ImageJ software, and the ratio of tetramer to dimer was estimated. *, P < 0.05; otherwise, P > 0.05.
DISCUSSION
The paramyxovirus fusion machine consists of an attachment glycoprotein protein that attaches the virus to host cell and a fusion protein that mediates membrane mixing. Receptor binding by the HN protein is believed to initiate the signal for fusion (15, 20, 50). However, recent studies have shown that receptor binding is dispensable for cell-cell fusion (2, 17, 19, 20, 43, 48). It is thought that the distal FAR of the HN1-117 stalk interacts constitutively with the Ig-like domain of F. This results in enhanced fusion compared with full-length HN (17, 20). Thus, HN1-117 is more than a mere anchor for the bulky receptor binding head. Some evidence suggests that the stability and conformational flexibility of the stalk play a regulatory role in HN/G- or H-mediated activation of their cognate F proteins (43). Also, changes in the length of HN1-117 abolish fusion promotion (see Table S1 in reference 17), suggesting a role in fine-tuning F activation. The atomic structure of NDV HN and PIV5 HN revealed a dimer of dimers and a tetramer, respectively, and a 4HB stalk (18, 32), but each of these two structural models lacks electron density for the MPSR portion of the stalk. Previous mutations introduced into the MPSR of PIV5 HN were found to regulate protein turnover and internalization (45), but a direct role in fusion promotion has not been explored. In this report, we investigated factors that influence HN1-117 stability and fusion. Specifically, we probed how the structurally unresolved MPSR influences HN1-117 stability and biological functions.
We assessed the role of the MPSR in structure-function studies of HN1-117 and full-length HN. First, triple consecutive (3×) Ala mutants were introduced into the MPSR of HN1-117 to examine its role in fusion promotion. We found that all but two of the 3× Ala mutants, consisting of residues 40ITQKQI45, promoted fusion (Fig. 1C and D and 2A), although to various degrees. The inhibition of fusion found in mutants MPSR2 and MPSR3 was not largely due to defects in protein expression or stability but correlates with changes in glycosylation of HN1-117 that may influence or be associated with the conformation and fusion promotion function consistent with previous findings (34, 51, 52). The change in mobility of MPSR2 and MPSR3 species on gels suggests that the glycosylation process of the HN1-117 differed for fusion-defective mutants from that of WT HN1-117 (Fig. 1C). HN1-117 T41, Q42, and I45 mutants were fusion inactive, while proteins with the same mutations in full-length HN were not (Fig. 4). The lack of fusion promotion is not due to defects in processing or protein transport (Fig. 3C). The altered mobility on gels (Fig. 3C) is suggestive of changes in glycosylation of HN1-117, which could affect the fusion-permissive conformation of the 4HB, thus disfavoring HN stalk-mediated F triggering. Another possible explanation is that the headless HN1-117 4HB conformation is vulnerable to the least modification. For example, these mutations may alter the FAR orientation such that the F interaction becomes disfavored. The lack of conformational stability in the absence of the receptor head is not unreasonable given that recently headless morbillivirus H was found to require stabilization for proper function (43).
We also examined the effect of the same mutations in the full-length HN. The single Ala mutations within the MPSR did not alter HN intracellular protein expression levels as well cell surface expression levels (Fig. 4A and B). However, receptor binding and NA activity were significantly increased (Fig. 4C and D). It can be inferred from these data that the primary sequence of the HN MPSR plays an important role in cell surface receptor binding and NA activity without influencing F protein triggering (Fig. 4E). These differences are reconcilable by the fact that the presence of the bulky head may stabilize the stalk by enhancing tight packing of the four stalks. The structural and mechanistic explanation for the severalfold increase in receptor binding and NA activity is not clear. One possible explanation is that the primary sequence at the MPSR specifies interhelical interactions that restrict the movements of each head subunit or an effect on the dimer-tetramer stability, which could impact HAd and NA activities. Hence, changing the stalk residues relieves this inhibitory mechanism and allows the HN head to adopt a conformation favoring enhanced HAd and NA activities.
To test this idea, cysteine mutations were introduced at residues 40 to 45 of HN in an attempt to trap the stalk in a disulfide-liked tetramer and to limit the degree of movement. Disulfide-linked stabilization of the MPSR of HN enhanced protein expression and cell surface expression (Fig. 5A and B). However, stabilization via disulfide-linked tetramers of the MPSR resulted in a modest increase in the receptor binding activity of HN (compared with corresponding Ala mutations [Fig. 4]), while reduction of the disulfide bond with TCEP yielded residue-specific increases in HAd (Fig. 5C). Recently, a number of mutations in the head-proximal region of the PIV5 and NDV HN1-117 that influence HAd, NA activity, and fusion have been described (18). Mutations in NDV HN1-117 (residues A89, L90, and L94) were found to inhibit fusion promotion with no adverse effect on receptor binding, NA activity, or antigenic recognition (53). Taken together, it can be inferred that multiple forces, including the stability of the 4HB and the degree of inter- and intraprotomer movement, regulate receptor recognition and NA activity, as well as F triggering by the HN protein.
Recently, biochemical evidence indicated that the MPSR of measles virus H protein adopts a structure distinct from the central and upper part of the stalk which forms a 4HB (54). Using Cys mutagenesis and oxidative cross-linking, we examined the formation of disulfide-linked tetramers at the MPSR in both HN1-117 and full-length HN. The formation of disulfide-linked tetramers within the MPSR did not enhance cell surface expression of HN1-117 (Fig. 6B). Oxidative cross-linking of the 4HB with CuP resulted in only a few of the Cys mutants with a tetramer/dimer ratio of >1. However, oxidative cross-linking of Cys residues at the MPSR of the full-length HN showed increased trapping of the 4HB (Fig. 7) with a tetramer/dimer ratio of >1, which is consistent with a more stable tetramer. Formation of disulfide-linked tetramers of the HN was accompanied by increased cell expression and surface export. Overall, it can be deduced that the presence of the head group enhances the oligomerization of HN1-117 with MPSR residues close enough to form disulfide-linked tetramers (Fig. 7). The loosening of the 4HB stalk of the HN1-117 stalk may explain its susceptibility to conformational alterations that impact its fusion triggering (17).
In summary, the amino acid sequence of the PIV5 HN MPSR plays an important role in the function of the stalk. This role of the MPSR may be important in fine-tuning receptor binding and NA activity. The stalk appears to affect the oligomerization of the NA head group (15), and our data suggest that the receptor binding head reciprocally influences the stability of the HN stalk.
ACKNOWLEDGMENTS
This research was supported in part by National Institutes of Health Research grants R01 AI 23173 (to R.A.L.) and R01 GM 61050 (to T.S.J.). R.A.L. is an Investigator of the Howard Hughes Medical Institute. E.A.-G. was supported by the UNCF-Merck Postdoctoral Science Research Fellowship.
REFERENCES
- 1.Bissonnette ML, Donald JE, DeGrado WF, Jardetzky TS, Lamb RA. 2009. Functional analysis of the transmembrane domain in paramyxovirus F protein-mediated membrane fusion. J Mol Biol 386:14–36. doi: 10.1016/j.jmb.2008.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bose S, Heath CM, Shah PA, Alayyoubi M, Jardetzky TS, Lamb RA. 2013. Mutations in the parainfluenza virus 5 fusion protein reveal domains important for fusion triggering and metastability. J Virol 87:13520–13531. doi: 10.1128/JVI.02123-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dutch RE, Hagglund RN, Nagel MA, Paterson RG, Lamb RA. 2001. Paramyxovirus fusion (F) protein: a conformational change on cleavage activation. Virology 281:138–150. doi: 10.1006/viro.2000.0817. [DOI] [PubMed] [Google Scholar]
- 4.Dutch RE, Jardetzky TS, Lamb RA. 2000. Virus membrane fusion proteins: biological machines that undergo a metamorphosis. Biosci Rep 20:597–612. doi: 10.1023/A:1010467106305. [DOI] [PubMed] [Google Scholar]
- 5.Paterson RG, Hiebert SW, Lamb RA. 1985. Expression at the cell surface of biologically active fusion and hemagglutinin-neuraminidase proteins of the paramyxovirus simian virus 5 from cloned cDNA. Proc Natl Acad Sci U S A 82:7520–7524. doi: 10.1073/pnas.82.22.7520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Horvath CM, Paterson RG, Shaughnessy MA, Wood R, Lamb RA. 1992. Biological activity of paramyxovirus fusion proteins: factors influencing formation of syncytia. J Virol 66:4564–4569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Doms RW, Lamb RA, Rose JK, Helenius A. 1993. Folding and assembly of viral membrane proteins. Virology 193:545–562. doi: 10.1006/viro.1993.1164. [DOI] [PubMed] [Google Scholar]
- 8.Bagai S, Lamb RA. 1995. Individual roles of N-linked oligosaccharide chains in intracellular transport of the paramyxovirus SV5 fusion protein. Virology 209:250–256. doi: 10.1006/viro.1995.1251. [DOI] [PubMed] [Google Scholar]
- 9.Paterson RG, Johnson ML, Lamb RA. 1997. Paramyxovirus fusion (F) protein and hemagglutinin-neuraminidase (HN) protein interactions: intracellular retention of F and HN does not affect transport of the homotypic HN or F protein. Virology 237:1–9. doi: 10.1006/viro.1997.8759. [DOI] [PubMed] [Google Scholar]
- 10.Melikyan GB, Jin H, Lamb RA, Cohen FS. 1997. The role of the cytoplasmic tail region of influenza virus hemagglutinin in formation and growth of fusion pores. Virology 235:118–128. doi: 10.1006/viro.1997.8686. [DOI] [PubMed] [Google Scholar]
- 11.Waning DL, Russell CJ, Jardetzky TS, Lamb RA. 2004. Activation of a paramyxovirus fusion protein is modulated by inside-out signaling from the cytoplasmic tail. Proc Natl Acad Sci U S A 101:9217–9222. doi: 10.1073/pnas.0403339101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yin HS, Wen X, Paterson RG, Lamb RA, Jardetzky TS. 2006. Structure of the parainfluenza virus 5 F protein in its metastable, prefusion conformation. Nature 439:38–44. doi: 10.1038/nature04322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Swanson K, Wen X, Leser GP, Paterson RG, Lamb RA, Jardetzky TS. 2010. Structure of the Newcastle disease virus F protein in the post-fusion conformation. Virology 402:372–379. doi: 10.1016/j.virol.2010.03.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Takimoto T, Taylor GL, Connaris HC, Crennell SJ, Portner A. 2002. Role of the hemagglutinin-neuraminidase protein in the mechanism of paramyxovirus-cell membrane fusion. J Virol 76:13028–13033. doi: 10.1128/JVI.76.24.13028-13033.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yuan P, Thompson T, Wurzburg BA, Paterson RG, Lamb RA, Jardetzky TS. 2005. Structural studies of the parainfluenza virus 5 hemagglutinin-neuraminidase tetramer in complex with its receptor, sialyllactose. Structure 13:803–815. doi: 10.1016/j.str.2005.02.019. [DOI] [PubMed] [Google Scholar]
- 16.Bose S, Welch BD, Kors CA, Yuan P, Jardetzky TS, Lamb RA. 2011. Structure and mutagenesis of the parainfluenza virus 5 hemagglutinin-neuraminidase stalk domain reveals a four-helix bundle and the role of the stalk in fusion promotion. J Virol 85:12855–12866. doi: 10.1128/JVI.06350-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bose S, Zokarkar A, Welch BD, Leser GP, Jardetzky TS, Lamb RA. 2012. Fusion activation by a headless parainfluenza virus 5 hemagglutinin-neuraminidase stalk suggests a modular mechanism for triggering. Proc Natl Acad Sci U S A 109:E2625–E2634. doi: 10.1073/pnas.1213813109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Welch BD, Yuan P, Bose S, Kors CA, Lamb RA, Jardetzky TS. 2013. Structure of the parainfluenza virus 5 (PIV5) hemagglutinin-neuraminidase (HN) ectodomain. PLoS Pathog 9:e1003534. doi: 10.1371/journal.ppat.1003534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jardetzky TS, Lamb RA. 2014. Activation of paramyxovirus membrane fusion and virus entry. Curr Opin Virol 5:24–33. doi: 10.1016/j.coviro.2014.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bose S, Song AS, Jardetzky TS, Lamb RA. 2014. Fusion activation through attachment protein stalk domains indicates a conserved core mechanism of paramyxovirus entry into cells. J Virol 88:3925–3941. doi: 10.1128/JVI.03741-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Poor TA, Jones LM, Sood A, Leser GP, Plasencia MD, Rempel DL, Jardetzky TS, Woods RJ, Gross ML, Lamb RA. 2014. Probing the paramyxovirus fusion (F) protein-refolding event from pre- to postfusion by oxidative footprinting. Proc Natl Acad Sci U S A 111:E2596–E2605. doi: 10.1073/pnas.1408983111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Russell CJ, Jardetzky TS, Lamb RA. 2004. Conserved glycine residues in the fusion peptide of the paramyxovirus fusion protein regulate activation of the native state. J Virol 78:13727–13742. doi: 10.1128/JVI.78.24.13727-13742.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tsurudome M, Ito M, Nishio M, Nakahashi M, Kawano M, Komada H, Nosaka T, Ito Y. 2011. Identification of domains on the fusion (F) protein trimer that influence the hemagglutinin-neuraminidase specificity of the F protein in mediating cell-cell fusion. J Virol 85:3153–3161. doi: 10.1128/JVI.01666-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zokarkar A, Lamb RA. 2012. The paramyxovirus fusion protein C-terminal region: mutagenesis indicates an indivisible protein unit. J Virol 86:2600–2609. doi: 10.1128/JVI.06546-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Russell CJ, Jardetzky TS, Lamb RA. 2001. Membrane fusion machines of paramyxoviruses: capture of intermediates of fusion. EMBO J 20:4024–4034. doi: 10.1093/emboj/20.15.4024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hiebert SW, Paterson RG, Lamb RA. 1985. Hemagglutinin-neuraminidase protein of the paramyxovirus simian virus 5: nucleotide sequence of the mRNA predicts an N-terminal membrane anchor. J Virol 54:1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Paterson RG, Hiebert SW, Lamb RA. 1985. Primary structure of the fusion protein and the hemagglutinin-neuraminidase protein of the paramyxovirus SV5, p 303–308. In Chanock RM, Lerner RA (ed), Modern approaches to vaccines: molecular and chemical basis of virus virulence and immunogenicity. Cold Spring Harbor Publications, Cold Spring Harbor, NY. [Google Scholar]
- 28.Porotto M, Fornabaio M, Greengard O, Murrell MT, Kellogg GE, Moscona A. 2006. Paramyxovirus receptor-binding molecules: engagement of one site on the hemagglutinin-neuraminidase protein modulates activity at the second site. J Virol 80:1204–1213. doi: 10.1128/JVI.80.3.1204-1213.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schmitt AP, He B, Lamb RA. 1999. Involvement of the cytoplasmic domain of the hemagglutinin-neuraminidase protein in assembly of the paramyxovirus simian virus 5. J Virol 73:8703–8712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schmitt AP, Leser GP, Waning DL, Lamb RA. 2002. Requirements for budding of paramyxovirus simian virus 5 virus-like particles. J Virol 76:3952–3964. doi: 10.1128/JVI.76.8.3952-3964.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Waning DL, Schmitt AP, Leser GP, Lamb RA. 2002. Roles for the cytoplasmic tails of the fusion and hemagglutinin-neuraminidase proteins in budding of the paramyxovirus simian virus 5. J Virol 76:9284–9297. doi: 10.1128/JVI.76.18.9284-9297.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yuan P, Swanson KA, Leser GP, Paterson RG, Lamb RA, Jardetzky TS. 2011. Structure of the Newcastle disease virus hemagglutinin-neuraminidase (HN) ectodomain reveals a four-helix bundle stalk. Proc Nat Acad Sci U S A 108:14920–14925. doi: 10.1073/pnas.1111691108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bose S, Zokarkar A, Welch BD, Leser GP, Jardetzky TS, Lamb RA. 2012. Fusion activation by a headless parainfluenza virus 5 hemagglutinin-neuraminidase stalk suggests a modular mechanism for triggering. Proc Natl Acad Sci U S A 109:E2625–2634. doi: 10.1073/pnas.1213813109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Melanson VR, Iorio RM. 2006. Addition of N-glycans in the stalk of the Newcastle disease virus HN protein blocks its interaction with the F protein and prevents fusion. J Virol 80:623–633. doi: 10.1128/JVI.80.2.623-633.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Apte-Sengupta S, Negi S, Leonard VH, Oezguen N, Navaratnarajah CK, Braun W, Cattaneo R. 2012. Base of the measles virus fusion trimer head receives the signal that triggers membrane fusion. J Biol Chem 287:33026–33035. doi: 10.1074/jbc.M112.373308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mattera R, Farías GG, Mardones GA, Bonifacino JS. 2014. Co-assembly of viral envelope glycoproteins regulates their polarized sorting in neurons. PLoS Pathog 10:e1004107. doi: 10.1371/journal.ppat.1004107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Malvoisin E, Wild TF. 1993. Measles virus glycoproteins: studies on the structure and interaction of the haemagglutinin and fusion proteins. J Gen Virol 74:2365–2372. doi: 10.1099/0022-1317-74-11-2365. [DOI] [PubMed] [Google Scholar]
- 38.Lee JK, Prussia A, Paal T, White LK, Snyder JP, Plemper RK. 2008. Functional interaction between paramyxovirus fusion and attachment proteins. J Biol Chem 283:16561–16572. doi: 10.1074/jbc.M801018200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Plemper RK, Hammond AL, Cattaneo R. 2001. Measles virus envelope glycoproteins hetero-oligomerize in the endoplasmic reticulum. J Biol Chem 276:44239–44246. doi: 10.1074/jbc.M105967200. [DOI] [PubMed] [Google Scholar]
- 40.Feldman SA, Crim RL, Audet SA, Beeler JA. 2001. Human respiratory syncytial virus surface glycoproteins F, G and SH form an oligomeric complex. Arch Virol 146:2369–2383. doi: 10.1007/s007050170009. [DOI] [PubMed] [Google Scholar]
- 41.Hu X, Ray R, Compans RW. 1992. Functional interactions between the fusion protein and hemagglutinin-neuraminidase of human parainfluenza viruses. J Virol 66:1528–1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Plemper RK, Brindley MA, Iorio RM. 2011. Structural and mechanistic studies of measles virus illuminate paramyxovirus entry. PLoS Pathog 7:e1002058. doi: 10.1371/journal.ppat.1002058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brindley MA, Suter R, Schestak I, Kiss G, Wright ER, Plemper RK. 2013. A stabilized headless measles virus attachment protein stalk efficiently triggers membrane fusion. J Virol 87:11693–11703. doi: 10.1128/JVI.01945-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yuan P, Leser GP, Demeler B, Lamb RA, Jardetzky TS. 2008. Domain architecture and oligomerization properties of the paramyxovirus PIV 5 hemagglutinin-neuraminidase (HN) protein. Virology 378:282–291. doi: 10.1016/j.virol.2008.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Leser GP, Ector KJ, Ng DTW, Shaughnessy MA, Lamb RA. 1999. The signal for clathrin-mediated endocytosis of the paramyxovirus SV5 HN protein resides at the transmembrane domain-ectodomain boundary region. Virology 262:79–92. doi: 10.1006/viro.1999.9890. [DOI] [PubMed] [Google Scholar]
- 46.Bauer CM, Pinto LH, Cross TA, Lamb RA. 1999. The influenza virus M2 ion channel protein: probing the structure of the transmembrane domain in intact cells by using engineered disulfide cross-linking. Virology 254:196–209. doi: 10.1006/viro.1998.9552. [DOI] [PubMed] [Google Scholar]
- 47.Liu Q, Stone JA, Bradel-Tretheway B, Dabundo J, Benavides Montano JA, Santos-Montanez J, Biering SB, Nicola AV, Iorio RM, Lu X, Aguilar HC. 2013. Unraveling a three-step spatiotemporal mechanism of triggering of receptor-induced Nipah virus fusion and cell entry. PLoS Pathog 9:e1003770. doi: 10.1371/journal.ppat.1003770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Brindley MA, Chaudhury S, Plemper RK. 2015. Measles virus glycoprotein complexes preassemble intracellularly and relax during transport to the cell surface in preparation for fusion. J Virol 89:1230–1241. doi: 10.1128/JVI.02754-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pinto LH, Dieckmann GR, Gandhi CS, Papworth CG, Braman J, Shaughnessy MA, Lear JD, Lamb RA, DeGrado WF. 1997. A functionally defined model for the M2 proton channel of influenza A virus suggests a mechanism for its ion selectivity. Proc Natl Acad Sci U S A 94:11301–11306. doi: 10.1073/pnas.94.21.11301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Villar E, Barroso IM. 2006. Role of sialic acid-containing molecules in paramyxovirus entry into the host cell: a minireview. Glycoconj J 23:5–17. doi: 10.1007/s10719-006-5433-0. [DOI] [PubMed] [Google Scholar]
- 51.Aguilar HC, Matreyek KA, Filone CM, Hashimi ST, Levroney EL, Negrete OA, Betrolotti-Ciarlet A, Choi DY, McHardy I, Fulcher JA, Su SV, Wolf MC, Kohatsu L, Baum LG, Lee B. 2006. N-glycans on Nipah virus fusion protein protect against neutralization but reduce membrane fusion and viral entry. J Virol 80:4878–4889. doi: 10.1128/JVI.80.10.4878-4889.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stone JA, Nicola AV, Baum LG, Aguilar HC. 2016. Multiple novel functions of henipavirus O-glycans: the first O-glycan functions identified in the paramyxovirus family. PLoS Pathog 12:e1005445. doi: 10.1371/journal.ppat.1005445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Melanson VR, Iorio RM. 2004. Amino acid substitutions in the F-specific domain in the stalk of the Newcastle disease virus HN protein modulate fusion and interfere with its interaction with the F protein. J Virol 78:13053–13061. doi: 10.1128/JVI.78.23.13053-13061.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Navaratnarajah CK, Kumar S, Generous A, Apte-Sengupta S, Mateo M, Cattaneo R. 2014. The measles virus hemagglutinin stalk: structures and functions of the central fusion activation and membrane-proximal segments. J Virol 88:6158–6167. doi: 10.1128/JVI.02846-13. [DOI] [PMC free article] [PubMed] [Google Scholar]