FIG 2.
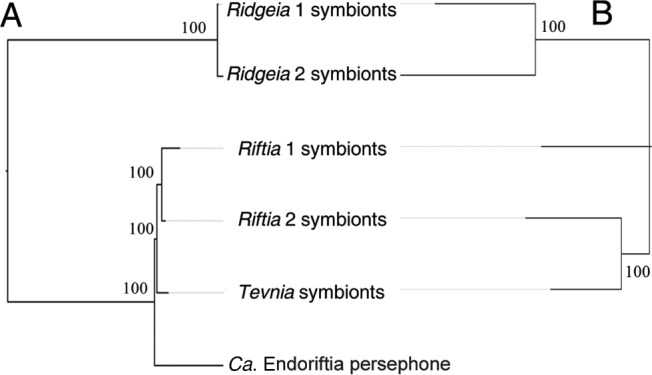
Neighbor-joining trees of “Candidatus Endoriftia persephone” based on the genetic distances (HKY model) between nucleotide sequences of the core genome (A) and the presence or absence of sequences of the accessory genome (B). (A) The six assemblies were aligned with Mauve and the locally colinear blocks extracted. Of these, only the LCBs of >100 bp that were represented in all assemblies were kept. The sequences within each LCB were aligned with MAAFT and were concatenated to form a genome-wide alignment of 2,580,528 bp containing 75,472 variable sites. (B) The first assembly of “Ca. Endoriftia persephone” (1) was not included in this analysis because of high genome fragmentation. Assemblies were aligned with Mauve, and the presence or absence of LCBs of >100 bp was used to generate a distance matrix (Jaccard index) from which a neighbor-joining tree was constructed using Populations, version 1.2.32. Bootstrap values are indicated at the branches.
