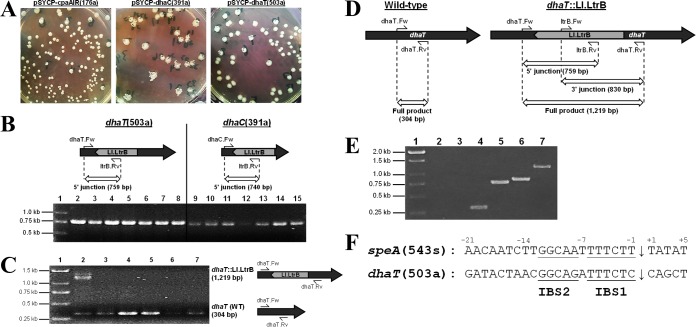
FIG 2.
Confirmation of mosaic and homogeneous intron disruption in C. pasteurianum dhaC and dhaT genes, respectively. (A) Colony morphology of transformants harboring intron donor constructs targeting sites cpaAIR(176a), dhaC(391a), and dhaT(503a). (B) Genomic verification of dhaT(503a) and dhaC(391a) intron insertion using one locus-specific primer and one intron-specific primer. Lane 1, marker; lanes 2 to 8, pSYCP-dhaT(503a) transformants; lanes 9 to 15, pSYCP-dhaC(391a) transformants. (C) Confirmation of mosaic intron insertion at site dhaT(503a) using locus-specific primers flanking the predicted intron insertion site. Lane 1, marker; lanes 2 to 7, pSYCP-dhaT(503a) transformants. The transformant screened in lane 2 shows a mosaic intron insertion. (D) Expected PCR products of the wild-type (left) and dhaT::Ll.LtrB mutant (right) strain. Insertion of the Ll.LtrB intron into the dhaT gene leads to a 915-bp increase in size of the full-length PCR product generated using primers flanking the 503a intron insertion site (dhaT.Fw and dhaT.Rv). Both 5′- and 3′-gene-intron junction PCR products can be detected in dhaT::Ll.LtrB mutant cells using primer sets dhaT.Fw and ltrB.Rv, and ltrB.Fw and dhaT.Rv, respectively. (E) Genomic verification of a single positive dhaT::Ll.LtrB mutant colony by amplification of all three PCR products depicted in Fig. 2A (5′ junction, 3′ junction, and full product). A wild-type C. pasteurianum colony was included as a control for all three PCR primer sets. Lane 1, marker; lanes 2 to 4, wild-type colony; lanes 5 to 7, dhaT::Ll.LtrB mutant colony; lanes 2 and 5, 5′ junction; lanes 3 and 6, 3′ junction; lanes 4 and 7, full product. (F) Comparison of chromosomal intron insertion sites corresponding to speA(543s) and dhaT(503a). The predicted intron insertion site is shown with an arrow. Sequence similarities in intron binding site 1 (IBS1) and IBS2 involved in intron base pairing with target DNA are underlined.