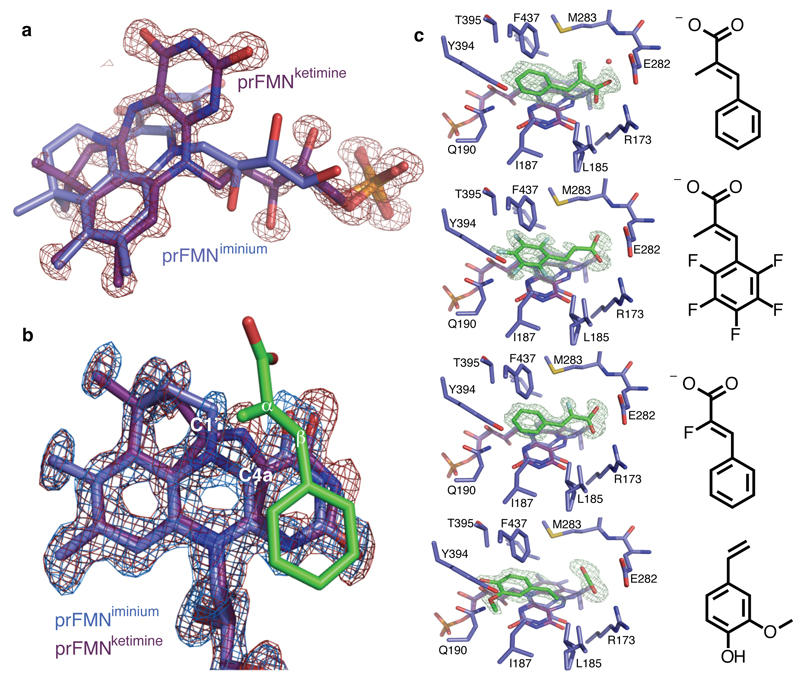
Fig 3. FdcUbiX cofactor structure and ligand complexes.
a) Omit electron density map corresponding to a distinct isomer of the prFMN contoured 5 sigma. An expansion of the central ring of the isoalloxazine system can clearly be observed, with the distinct butterfly-bent conformation accompanied by altered conformation of the ribityl moiety. b) Detailed view of prFMNox isoforms in the FdcUbiX-substrate complexes. The omit electron density maps corresponding respectively to the prFMNiminium (in blue) and prFMNketimine (in red) species each contoured 5 sigma are shown for the alpha-methyl-cinnamic acid complex. c) A series of FdcUbiX substrate/product complexes. Selected active site residues of FdcUbiX are shown in atom colour sticks, with the omit electron density contoured at 5 sigma corresponding to respectively alpha-methyl-cinnamic acid, pentafluorocinnamic acid, alpha-fluorocinnamic acid and 4-vinyl-guaiacol.