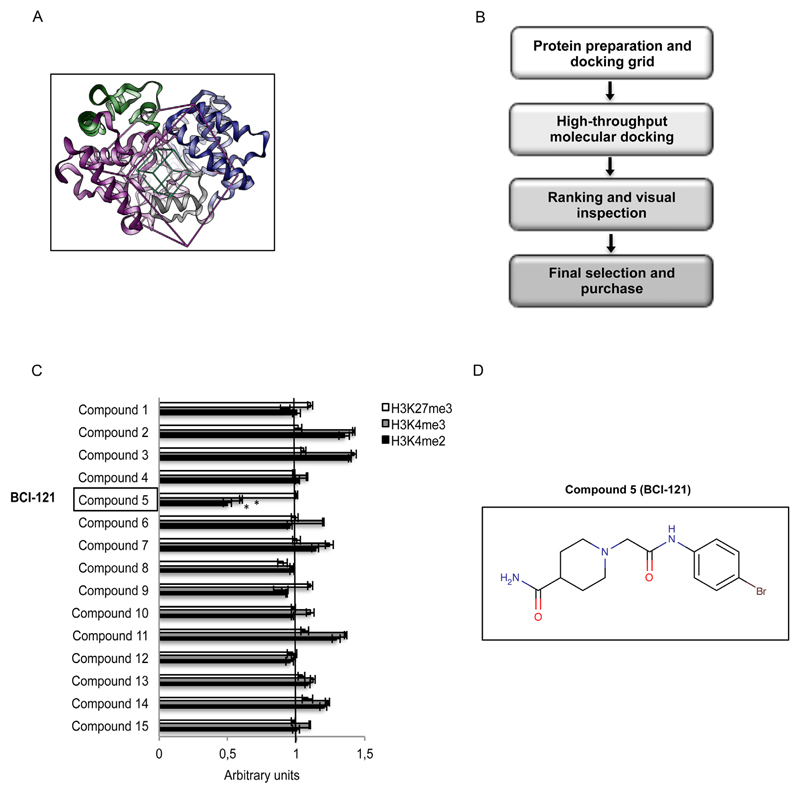
Fig. 4.
Virtual screening procedure allowing identification of BCI-121 as a promising candidate for SMYD3 inhibition. (A) Structure of full-length SMYD3 highlighting the N-terminal SET domain (purple), the MYND domain (yellow), the post-SET domain (gray), and the C-terminal region (blue). The boxes define the area of the protein - centered on the histone binding site - that was considered to perform the docking screening calculation. The green box identifies the area containing at least one atom of the putative ligand, while the purple box identifies the area where all atoms of the ligands should lie. (B) Schematic representation of the virtual screening procedure. (C) Compound 5 [BCI-121] is the best candidate small molecule for SMYD3 inhibition among the initial set of 15 compounds selected through the virtual screening procedure. The global level of SMYD3 targeted [H3K4me2 and H3K4me3] and non-targeted [H3K27me3] histone methyl marks was measured by immunoblot in nuclear enriched fractions of CRC cells (HT29) treated with each compound (100 μM). Values shown correspond to histone methyl mark levels quantified by densitometric analysis and normalized to the loading control Lamin A/C (arbitrary units, untreated control at 48h = 1). (D) Structural formula of the selected compound BCI-121 (Figure generated with MarvinSketch v6.0.0 http://www.chemaxon.com). Statistical analysis was performed using Student’s t-tail test; *P < 0.05 was considered statistically significant.