Figure 2.
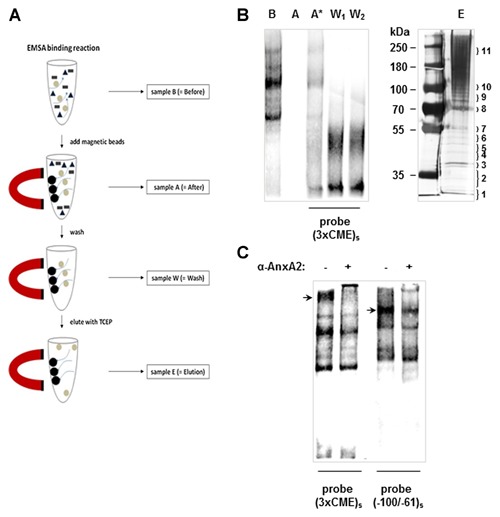
Isolation of CME binding proteins with Magnetic Bead Pull‐Down. The outline of the biotin‐streptavidin magnetic bead pull‐down assay is depicted in (A) with samples taken at various points for analysis in EMSA and SDS–PAGE as shown in (B). Briefly, SV‐WI38 nuclear proteins were incubated with the biotinylated single‐stranded (3xCME)s probe under optimised EMSA binding conditions. A sample was taken (B = Before) for EMSA analysis. Streptavidin‐coated magnetic beads were then added to remove the biotinlyated probe with bound protein complexes from solution by magnetic separation; a sample (A = After) was taken for EMSA analysis. To account for loss of probe and allow direct comparison of binding reactions, an appropriate amount of biotinylated probe was added in the EMSA (A* = After, with added probe). Protein complexes bound to the probe were washed twice and recovered by magnetic separation while the remaining supernatant was analysed by EMSA (W1 and W2 = Wash 1 and 2). Protein complexes were eluted from the probe with 250 mM TCEP and separated by SDS–PAGE with subsequent silver staining (E = elution). Numbers on the right correspond to the gel fragments that were cut out from the gel and subjected to MALDI–TOF mass spectrometry analysis. (C) EMSA showing blocking of complex formation to the (3xCME)s probe and the (−100/−61)s probe, respectively, when SV‐WI38 nuclear proteins were incubated in the presence of an anti‐AnxA2 antibody. Major protein complexes interacting with the antibody are indicated with an arrow.
