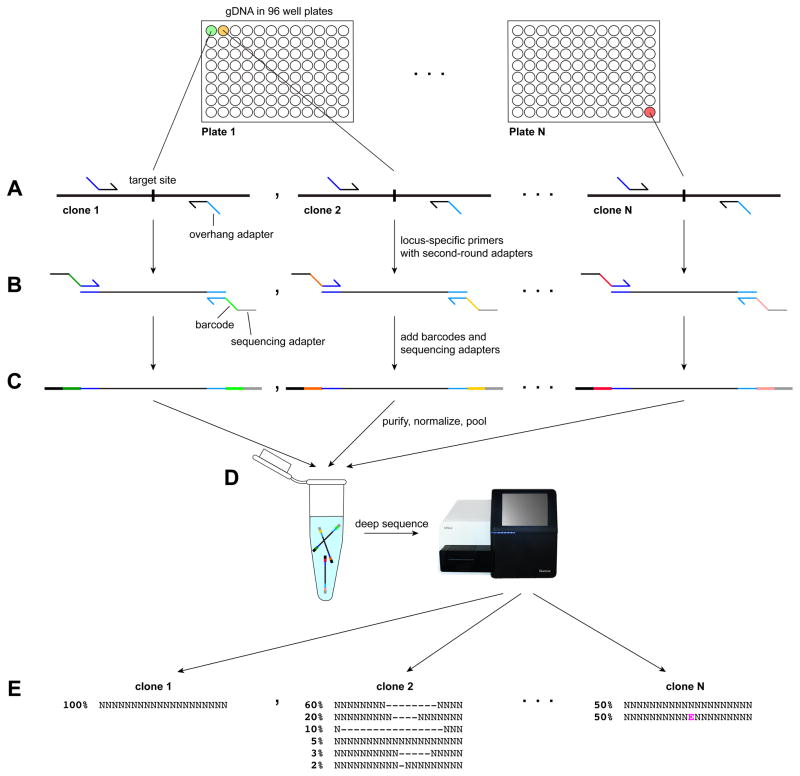
Figure 6.
Barcoded deep sequencing of gene-edited hPSC clones. A) Genomic DNA extracted from hPSCs is used as the template for PCRs to amplify the targeted region. These locus-specific PCR primers carry adapters that enable a second-round PCR. B) A brief second-round PCR adds unique barcodes and sequencing adapters to each amplicon. The schematic shown is for sequencing using dual-indexed, paired-end reads. C–D) Amplicons are gel-purified, concentration-normalized (C), and pooled into an amplicon library that can contain thousands of clones representing the same targeted locus or many different loci (D). PhiX is often spiked in to add diversity to the library and improve sequencing quality. E) Sequencing reads from each clone are aligned to a reference sequence and analyzed for mutations. In this example, clone 1 has no mutations and clone 2 is heterogeneous and would likely have to be subcloned. Clone N is an example of a successful heterozygous targeted editing experiment, in which 50% of the reads contain the desired single base change.