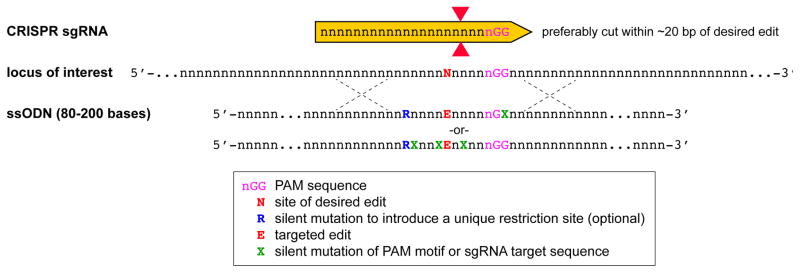
Figure 7.
Schematic for ssODN design. The CRISPR site should be designed so that the cut site (red triangles) is in close proximity to the site of the desired edit (red N). ssODNs should be designed to carry the desired edit with 40 to 100 bases of homologous sequence on either side. In addition, these ssODNs should carry silent substitutions (assuming a coding region is being targeted) that either mutates the PAM or the recognition domain so that the ssODN itself is not targeted and so that the modified locus is protected from further DSBs. In addition, it may be useful to introduce (or remove) a unique restriction site so that targeted clones can be screened by PCR and restriction digest as a backup method for sequencing. The sequences shown are for general reference and relative position of the sgRNA and the bases to be modified will vary depending on experimental needs.