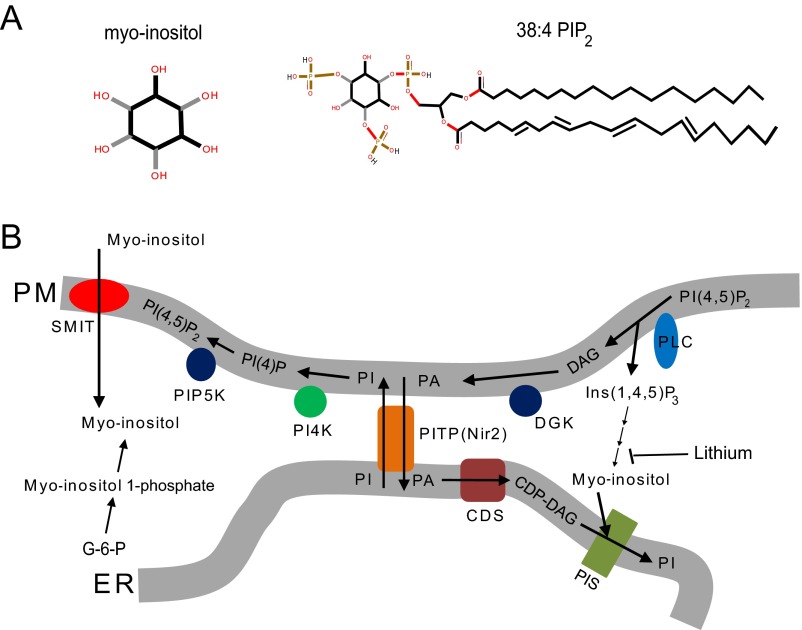
Fig. S1.
Related to Fig. 1. (A) Diagram showing the structure of myo-inositol and 38:4 PI(4,5)P2. The black and gray sticks in the myo-inositol structure indicate different geometrical orientations of the chemical bonds connecting the inositol ring and the hydroxyl group. (B) Diagram illustrating the metabolism of phosphoinositides (3). CDP-DAG, cytidine diphosphate diacylglycerol; CDS, CDP-DAG synthase; DAG, diacylglycerol; DGK, discylglycerol kinase; ER, endoplasmic reticulum; G-6-P, glucose-6-phosphate (glucose-6-phosphate is isomerized by the inositol-3-phosphate synthase to myo-inositol 1-phosphate); PA, phosphatidic acid; PI, phosphatidylinositol; PI4K, phosphatidylinositol 4-kinase; PI(4)P, phosphatidylinositol (4)-phosphate; PI(4,5)P2, phosphatidylinositol (4, 5)-biphosphate; PIP5K, phosphatidylinositol (4)-phosphate 5 kinase; PIS, phosphatidylinositol synthase; PITP, phosphatidylinositol transfer protein; PLC, phospholipase C; PM, plasma membrane; SMIT, sodium-coupled myo-inositol transporter. PITP(Nir2) can transfer PI from ER to PM and PA from PM to ER at ER–PM junctions (61).