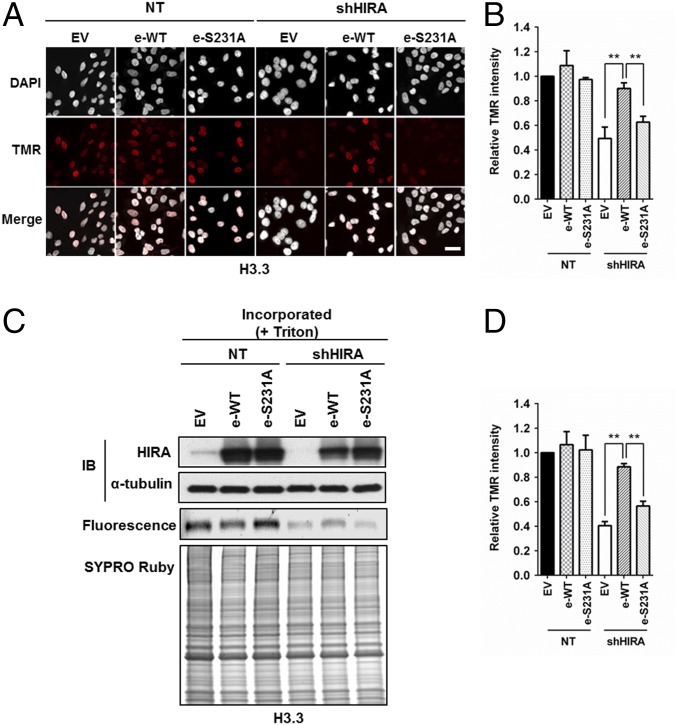
Fig. 4.
HIRA S231A mutant exhibits defects in de novo histone H3.3 deposition. (A and B) Exogenously expressed HIRA, but not HIRA S231A mutant, rescues the H3.3 defect caused by HIRA depletion. Endogenous HIRA was depleted from HeLa cells expressing H3.3-SNAP, and HIRA or HIRA S231A mutant was reexpressed. The effect of H3.3 deposition was monitored at individual cells by using microscopy. (A) Representative DAPI and H3.3-SNAP staining images. EV, empty vector. (Scale bar: 20 μm.) (B) Quantification of H3.3-SNAP staining intensity at individual cells (mean ± SEM, **P < 0.01, n = 3 biological replicates). (C and D) HIRA S231A mutant fails to complement H3.3 deposition defects as detected by chromatin fractionation assays. The experiments were performed as described above except that chromatin fractionation assays were used to monitor H3.3 deposition. Proteins in chromatin fractions were analyzed. C, Top shows analysis of HIRA depletion and expression of exogenous wild type HIRA (e-WT) or HIRA S231A mutant (e-S231A). H3.3-SNAP florescence was detected by phosphoImager (C, Middle) and total proteins by SYPRO Ruby staining (C, Bottom). Relative TMR intensity was calculated by normalizing H3.3-SNAP signals against whole protein levels (D, mean ± SEM, **P < 0.01, n = 3 biological replicates).