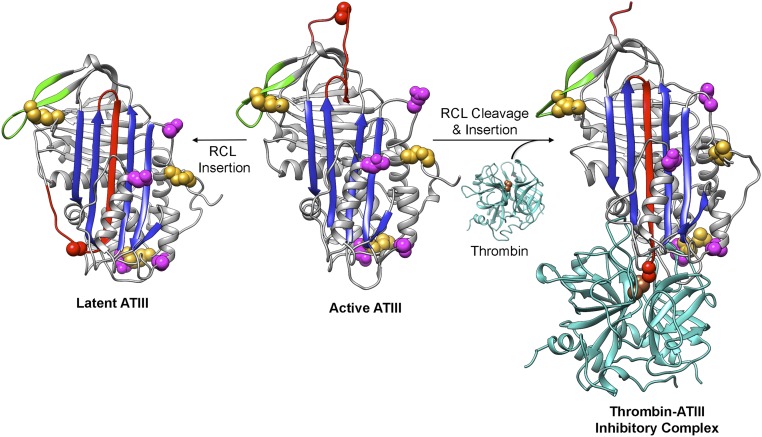
Fig. S1.
ATIII conformational changes. The RCL (red) can spontaneously insert into β-sheet A (the central sheet in blue), resulting in irreversibly inactivated latent ATIII. Cleavage of active, metastable ATIII by target proteases (cyan) at Arg393 (red space fill) leads to insertion of the N-terminal end of the cleaved RCL into sheet A, translocation of the protease relative to the serpin, and trapping of the serine protease acyl-enzyme intermediate with a covalent bond between ATIII Arg393 and thrombin Ser195 (brown space fill), the serine residue in the catalytic triad. Cys residues and the sites of N-linked glycosylation are shown as gold and magenta space filling representations, respectively. The active (heparin bound) and latent ATIII structures are from Protein Data Bank (PDB) ID code 1E05, and the thrombin structure is from PDB ID code 1PPB. The ATIII–thrombin inhibitory complex was modeled using the α1-antitrypsin–trypsin inhibitory complex structure (PDB ID code1EZX). The sequences of residues 3–393 of human ATIII (from Uniprot P01008) along with a 2.0 Å distance constraint for each of the N-terminal disulfide bonds were submitted to I-TASSER (37) (zhanglab.ccmb.med.umich.edu/I-TASSER/) along with the A chain of 1EZX as the template. Models of loop-inserted ATIII were generated, and the top model, the structure of the ATIII C-terminal residues 394–431 from PDB ID code 1E05, and the structure of α-thrombin (PDB ID code 1PPB) were structurally aligned with the α1-antitrypsin–trypsin inhibitory complex structure (PDB ID code 1EZX) using UCSF Chimera (38). The resulting model of the ATIII–thrombin inhibitory complex was then adjusted manually. All structure figures were made using UCSF Chimera (38).