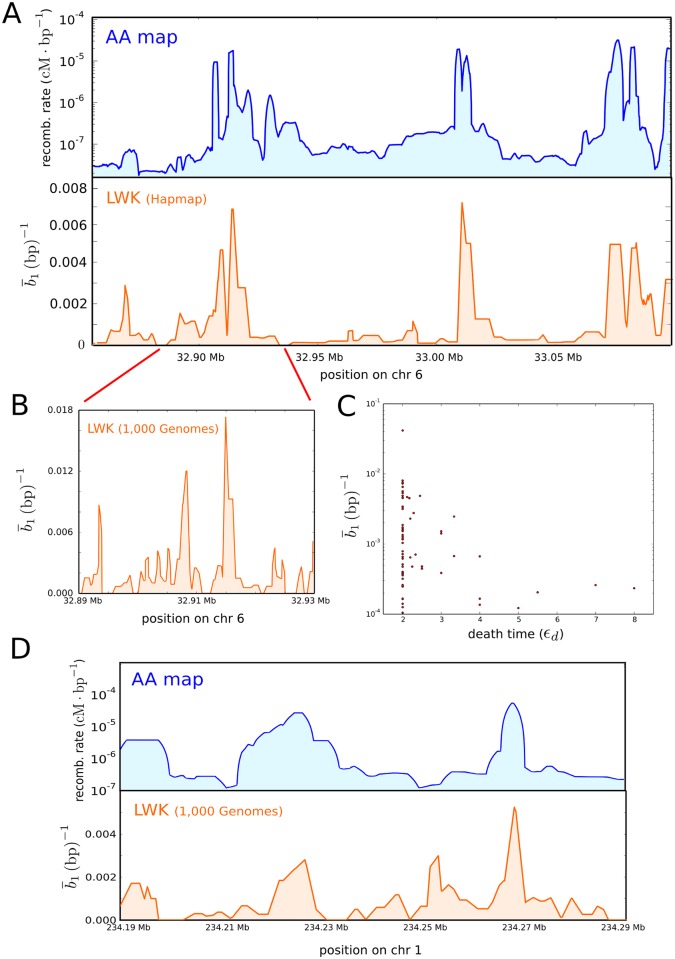
Fig 9. Barcode ensemble across the HLA and MS32 mini-satellite loci of the LWK population.
(A) Recombination rates (top) across a 250 kilobase region of the HLA locus according to the African-American recombination map, based on 30,000 individuals [55]. The vertical axis is in logarithmic scale. The distribution of recombination events (bottom) detected by the barcode ensemble of a sample of 90 individuals from the LWK population sequenced by the International HapMap Consortium [53] is consistent with the observed recombination rates. Note that in neutral models of evolution the number of recombination events in minimal ARGs is roughly expected to grow logarithmically with the recombination rate of the population [57]. (B) Distribution of recombination events detected by the barcode ensemble of a sample of 97 individuals from the LWK population, sequenced by the 1,000 Genomes Project Consortium [56]. The higher density of SNPs in this dataset allows for a higher resolution in the localization of recombination events as well as a higher sensitivity. (C) Density of recombination events per nucleotide against their average death time 2ϵd, for recombination events captured by the barcode ensemble in (A). Each point represents a genomic position for which the barcode ensemble detects recombination. The horizontal axis represents the average death time of the bars in the barcode ensemble that are associated to that genomic position. Events with large ϵd, corresponding to recombination events with a large mutational distance between recombining sequences, are mostly associated to regions with low number of recombinations, as expected from neutral models of evolution [57]. (D) Recombination rates (top) across a 100 kilobase region near the MS32 mini-satellite locus according to the African-American recombination map [55]. The vertical axis is in logarithmic scale. The distribution of recombination events (bottom) detected by the barcode ensemble of a sample of 97 individuals from the LWK populations sequenced by the 1,000 Genomes Project Consortium [56] is consistent with the observed recombination rates.