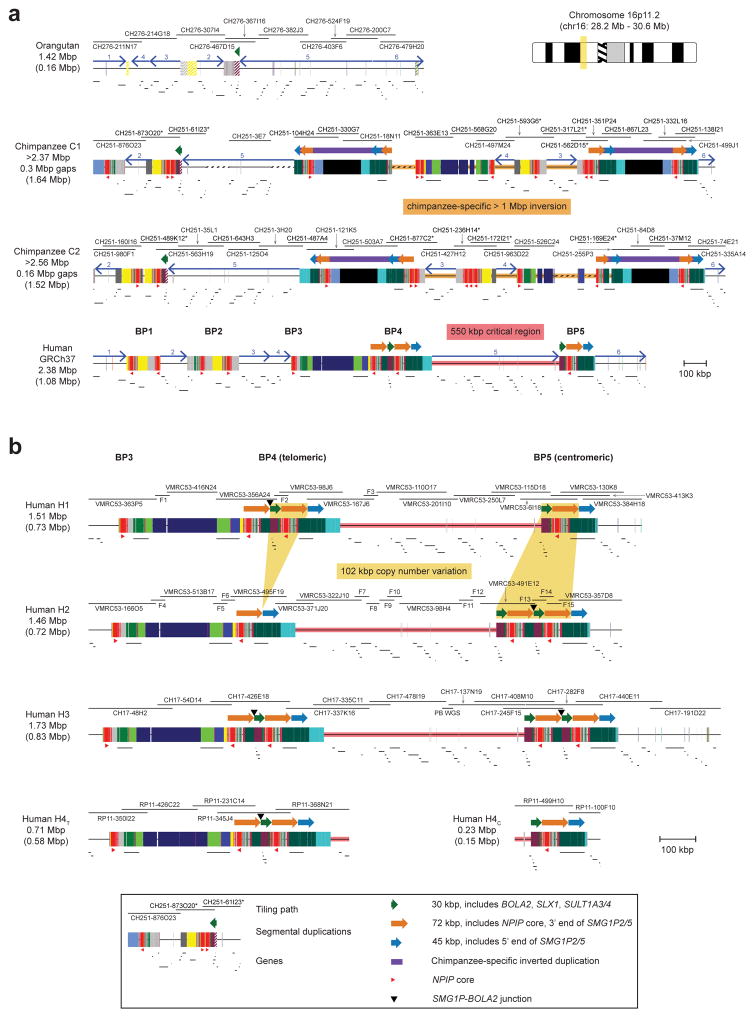
Extended Data Figure 1. Comparative sequence analysis of chromosome 16p11.2 among apes.
a) Schematic depicts the genomic organization of chromosome 16p11.2 for one orangutan and two chimpanzee haplotypes along with the human reference haplotype (GRCh37 chr16:28195661–30573128; see ideogram for approximate chromosomal location). Blocks of segmental duplications within this locus mediate recurrent rearrangements in humans; thus, these blocks have been defined as breakpoint regions BP1–BP5 (ref. 8). The ~550 kbp critical region (pink) and a >1 Mbp chimpanzee-specific inversion polymorphism (orange) are highlighted. Tiling paths of sequenced clones are indicated above each haplotype, with chimpanzee clones that could not be fully resolved marked with asterisks. Colored boxes and thick arrows indicate the extent and orientation of segmental duplications (with different colors denoting duplicons from different ancestral genomic loci, and hashed boxes indicating sequence duplicated in humans but not in the species represented). Thin numbered arrows show orientations of gene-rich regions of unique sequence. Numbers (left) indicate the size of each orthologous haplotype, with the number of segmentally duplicated base pairs shown in parentheses. Note that, for chimpanzee, these sizes are lower bounds due to gaps in the contigs (dotted line sections) and the contigs not reaching unique sequence beyond BP1 (i.e., unique region 1). b) Schematic depicts distinct human structural haplotypes over the chromosome 16p11.2 critical region and flanking sequences (three complete haplotypes extending from unique sequence distal to BP3 to unique sequence proximal to BP5 and one partial haplotype including BP3–BP4 and BP5 sequence contigs). High-quality sequence for each haplotype was generated by sequencing a total of 40 BACs and 15 fosmids from three different human genomic libraries. Regions of copy number variation (highlighted in yellow along the first two haplotypes) occur on both sides of the critical region and involve the same 102 kbp unit in direct orientation, including a 30 kbp block containing BOLA2 and two other genes and a 72 kbp block harboring a partial segmental duplication of SMG1 (SMG1P). Expansion and contraction of this cassette underlie hundreds of kbp of structural diversity between human haplotypes. BOLA2 paralog-specific copy number genotype data suggest that H1 and H3 likely represent the most common haplotype structures in humans.