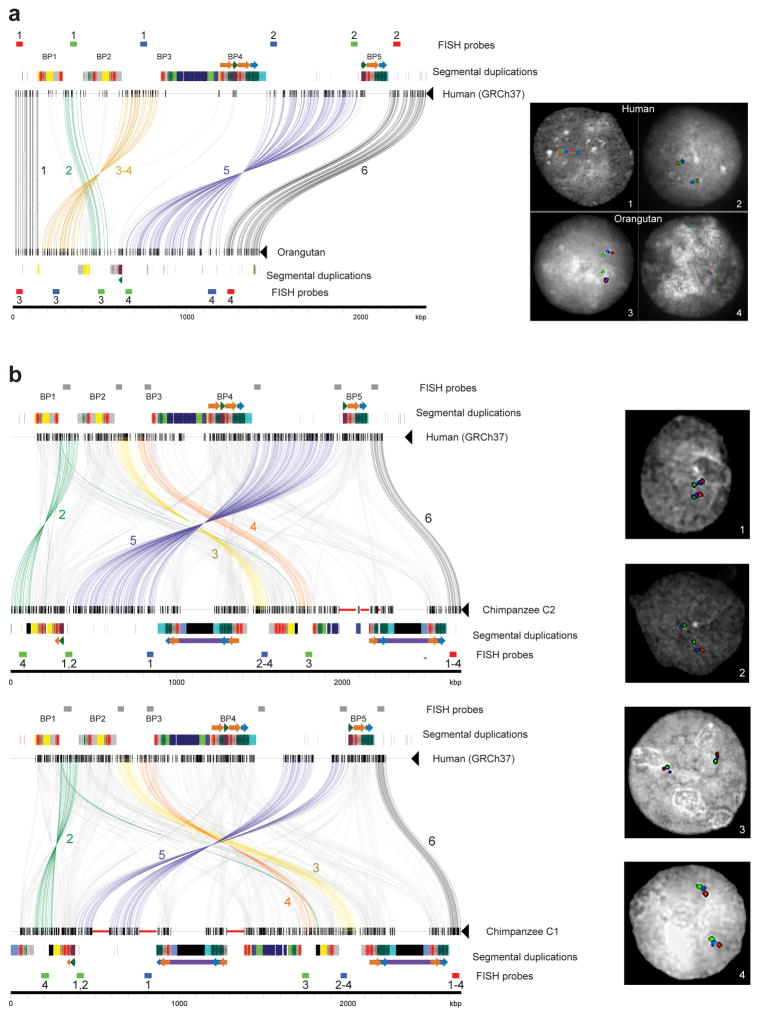
Extended Data Figure 2. Comparison of chromosome 16p11.2 structure between apes.
a) Sequences (thin horizontal lines) from human (GRCh37 chr16:28195661–30573128) and orangutan (contig sequence) at chromosome 16p11.2 are compared using Miropeats (s = 1,000) and annotated with locations of human segmental duplications and FISH probes used to validate the organization of the region. Lines connecting the sequences show regions of homology, and line colors highlight differences in the order and orientation of distinct gene-rich regions of unique sequence across the locus (numbered 1–6). Numbers below FISH probes correspond to numbers within the images on the right, specifying which probes were used in each experiment. Experiment 1 used the same probes as experiment 3, and experiment 2 used the same probes as experiment 4. Three-color interphase FISH on human and orangutan chromosomes confirms the accuracy of our assembled orangutan contig. b) Sequences (thin horizontal lines) from human (GRCh37 chr16:28195661–30573128) and two chimpanzee structural haplotypes at chromosome 16p11.2 are compared using Miropeats (s = 1,500) and annotated with locations of human segmental duplications and FISH probes used to validate the organization of the region. Thick red horizontal lines indicate gaps in the chimpanzee contigs, and black boxes correspond to chimpanzee-specific segmental duplications (i.e., sequences not duplicated in humans). Lines connecting the sequences show regions of homology, and line colors highlight differences in the order and orientation of distinct gene-rich regions of unique sequence across the locus (numbered 2–6). Numbers below FISH probes correspond to numbers within the images on the right, specifying which probes were used in each experiment. Gray rectangles show mapping locations of FISH probes in human. Three-color interphase FISH on chimpanzee chromosomes confirms the accuracy of our assembled contigs.