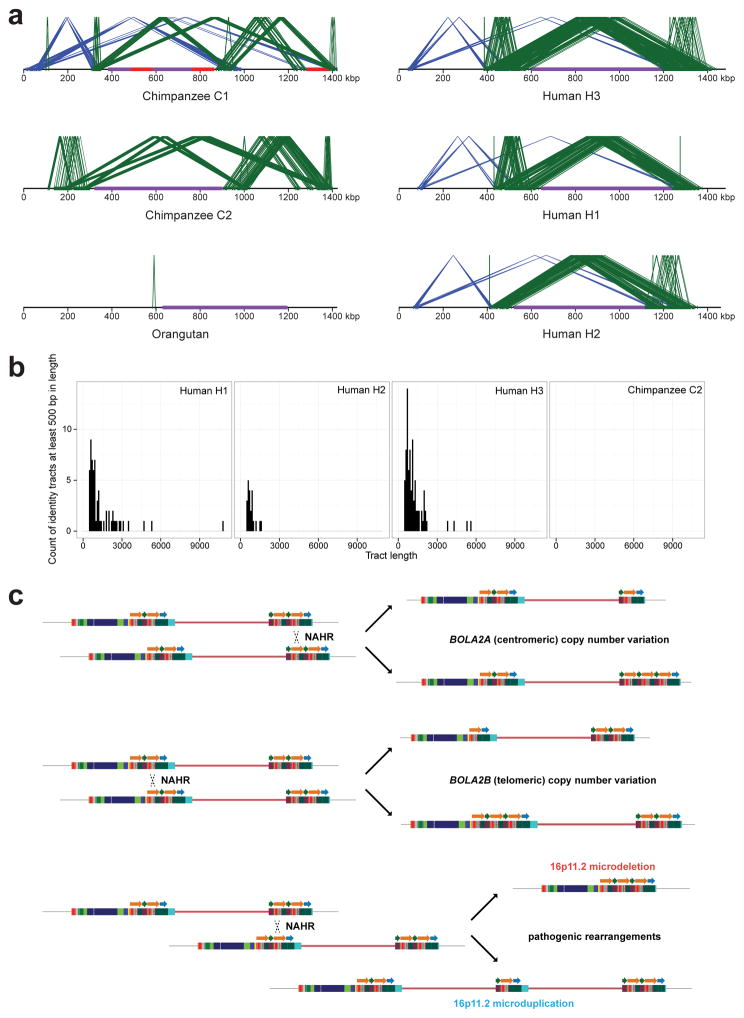
Extended Data Figure 5. Comparison of duplications around the chromosome 16p11.2 autism critical region among apes and NAHR model underlying CNV at human chromosome 16p11.2.
a) Local directly oriented (green) and inversely oriented (blue) intrachromosomal segmental duplications flanking the chromosome 16p11.2 autism critical region (purple) are visualized using Miropeats (s = 1,000). Gaps in the chimpanzee C1 contig are shown in red. The smaller size (<50 kbp) and lower average sequence identity (at most 98.6%) of directly oriented duplications flanking the critical region in chimpanzee compared to human haplotypes including BOLA2 on both sides of the critical region (at least 147 kbp of directly oriented duplications having at least 99.3% average sequence identity) suggest that susceptibility to NAHR resulting in microdeletions and microduplications at this locus evolved specifically in humans. b) Perfect sequence identity tract lengths (>500 bp) within directly oriented duplications flanking the critical region for human vs. chimpanzee. Histograms show counts of tracts of perfect sequence identity (lacking single-nucleotide variants and indels) between directly oriented segmental duplications of interest within each indicated haplotype and the distribution of these tracts over different size ranges. Human haplotypes having BOLA2 on both sides of the critical region (bottom panels) contain the highest number of such tracts and the longest such tracts, including one tract spanning 10,774 bp. In contrast, the longest tract of perfect sequence identity between duplications of interest in chimpanzee (considering both the C1 and C2 haplotypes) spans 450 bp. c) NAHR model underlying normal and disease-associated CNV at human chromosome 16p11.2.