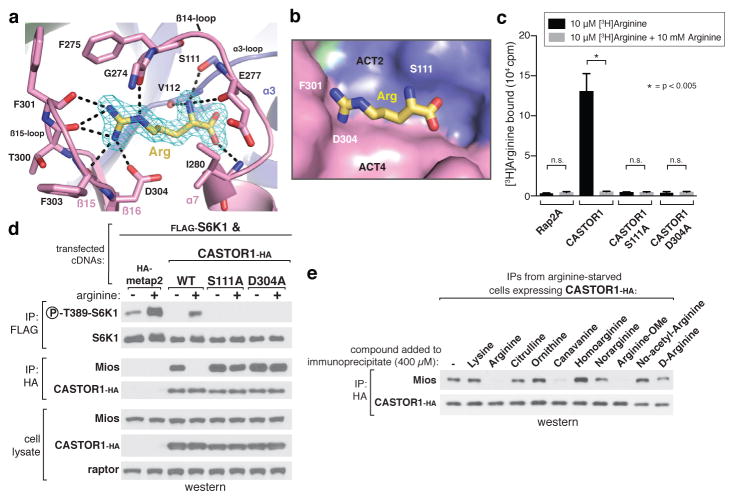
Figure 2. The arginine-binding pocket of CASTOR1.
a, View of the arginine-binding pocket in CASTOR1, together with its Fo-Fc electron density map calculated and contoured at 4σ from an omit map lacking arginine. The bound arginine is shown in yellow. Hydrogen bonds or salt-bridges are shown as black dashed lines. Residues 269–273 are omitted for clarity. b, Steric view of the arginine-binding pocket, depicting the surface representation of CASTOR1 and stick model of arginine (yellow). The β14-loop (residues 269–280) is omitted for clarity. c, CASTOR1 S111A and D304A mutants do not bind arginine in vitro. FLAG-immunoprecipitates prepared from HEK-293T cells transiently expressing indicated FLAG-tagged proteins were used in binding assays with [3H]Arginine as described in the methods. Values are Mean ± SD for 3 technical replicates from one representative experiment. d, The CASTOR1 S111A and D304A mutants constitutively bind GATOR2 and inhibit mTORC1 signaling in cells. HEK-293T cells transiently expressing FLAG-S6K1 and the indicated HA-tagged constructs were starved of arginine for 50 min and, where indicated, restimulated for 10 min. Both FLAG- and HA-immunoprecipitates were prepared from lysates and analyzed as in 1c. e, Effects of various arginine analogues on the CASTOR1-GATOR2 interaction in vitro. HEK-293T cells transiently expressing HA-CASTOR1 WT were starved of arginine for 50 min. HA-immunoprecipitates were prepared from cell lysates then incubated with 400 μM of the indicated compounds for 20 min and analyzed as in 1c.