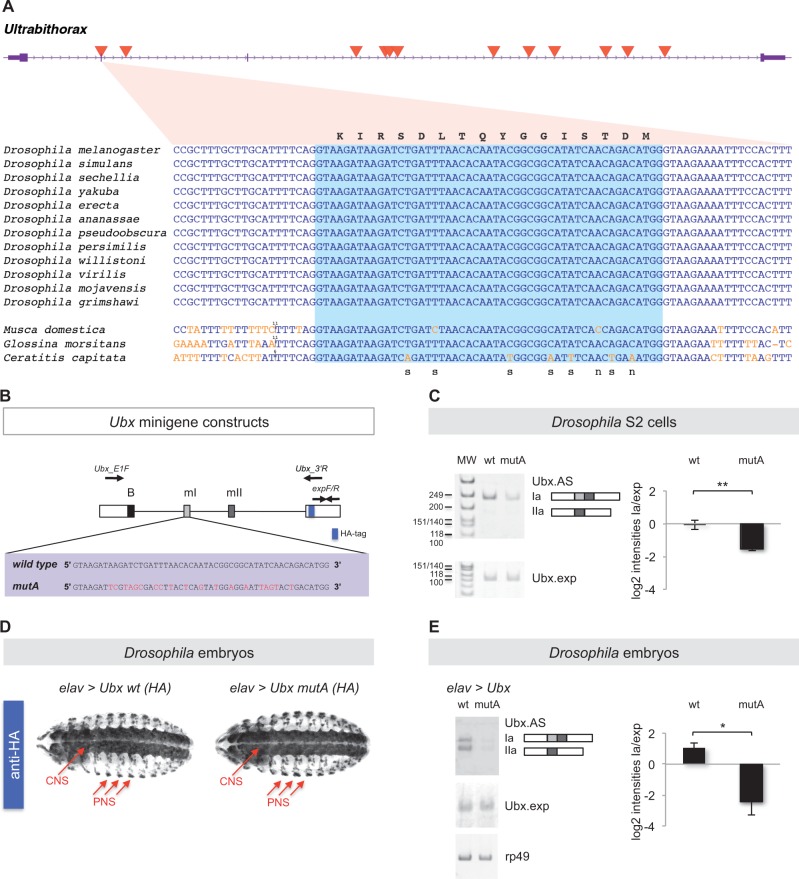
Fig. 4.
Analysis of an ultraconserved exon within the Ubx gene. (A) The Ubx gene (purple) contains 12 UCEs (red triangles) within the transcribed region. Coding sequences are shown as thick boxes and untranslated regions (UTRs) as narrow boxes. The gene is transcribed from left to right. One of the UCEs overlaps with the alternatively spliced mI exon. An alignment of the mI-UCE sequence from 12 Drosophila species and three more distantly related fly species is shown. Substitutions relative to the Drosophila sequence are shown in orange and insertions as a vertical line, with the number of inserted bases added above the sequence. The light blue box highlights the mI exon, while the surrounding sequences are intronic. The amino acid sequence (corresponding to the Drosophila nucleotide sequences) is shown above the alignment. For positions with observed substitutions, it is noted below the alignment whether these are synonymous (s) or nonsynonymous (n). (B) To explore the roles of microexon mI ultraconservation in Ubx splicing control we engineered a series of Ubx minigene constructs so that they included wild type (wt) or mutated versions of microexon mI (mutA) in which the protein coding potential of the gene was maintained while the ultraconserved nucleotide sequence of mI was disrupted by means of synonymous mutations (red). Approximate positions of splicing primers Ubx_E1F (forward) and Ubx_3′U (reverse) and expression primers expF/R (forward/reverse) are indicated. (C) Experiments in Drosophila Schneider 2 (S2) cells. Semi-quantitative RT-PCR analysis of wild type and mutA Ubx minigenes expressed in S2 cells reveals distinct patterns of Ubx mRNA splicing where the mutA minigene construct shows a marked reduction of Ubx Ia isoform production. Ubx.AS refers to the signal detected with primers Ubx_E1F and Ubx_3′R (see B) which detects all alternative splicing variants of the gene; Ubx.exp denotes signal amplified with primers expF/R (see B) which are positioned in the 3′ exon, a constitutive segment of Ubx mRNAs. (D) Expression of Ubx wild type and Ubx mutA minigenes in the Drosophila embryo. We produced HA-tagged UAS versions of wt and mutA Ubx minigenes (see A) and generated independent transgenic UAS-lines with insertions in identical chromosomal loci by means of site-specific recombination. The resulting UAS-Ubx lines (wt and mutA) were crossed with the elav-gal4 (elav) driver to express Ubx transgenes selectively within the developing embryonic nervous system. Note that expression patterns obtained with anti-HA antibodies in the embryonic CNS and PNS were identical across genotypes confirming comparable gene expression conditions. (E) Semi-quantitative RT-PCR analysis of wt and mutA Ubx minigene expression in the embryonic Drosophila nervous system reveals effects of mI on Ubx splicing control. In line with the results obtained in S2 cells (see C) we observed that the mutA minigene produced a reduced amount of Ubx Ia isoform when compared with its wild-type counterpart. (see C for definition of labels Ubx_AS and Ubx.exp and text for further details). Statistical analyses: **P < 0.01 and *P < 0.05 obtained in one-tailed t-test (P-value S2 cells = 0.0035 (**); P-value embryos = 0.0318 (*). Error bars indicate standard error of the mean. HA, haemagglutinin tag; B, Ubx B-element; mI, microexon I; mII, microexon II.