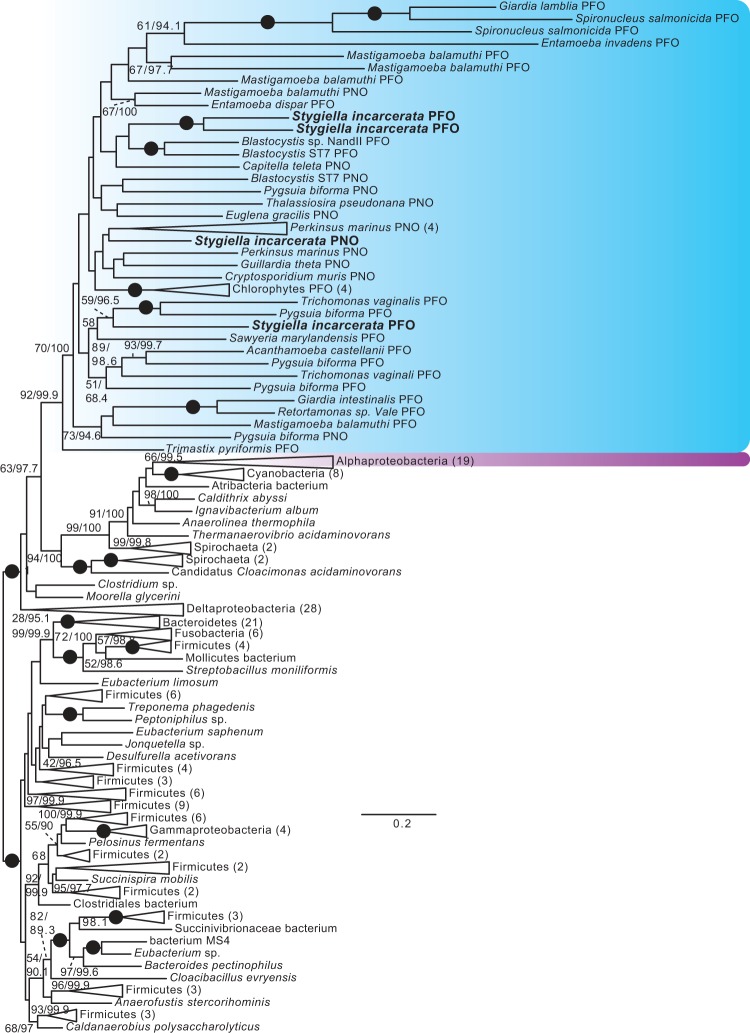
Fig. 4.
Unrooted ML tree of PFO sequences. Phylogenetic analyses were performed on 212 sequences and 958 sites, using RAxML v.8.0.23 under the PROTGAMMALG4X model of amino acid substitution, and IQ-TREE v.1.3.11 under the C20+Gamma model of evolution. Bootstrap support values >50% (first number), and ultrafast bootstrap values >95% (second number), are shown next to relevant branches; both support values are shown even where only one is greater than the chosen cutoff. Branches with 100% bootstrap and ultrafast bootstrap support are indicated by black circles. Eukaryotes are shaded blue, and alphaproteobacteria magenta. Eukaryotic PFO and PNO sequences are indicated. For ultrafast bootstrap values, 95 was chosen as a cutoff value as per the recommendation of the IQ-TREE manual; for the ML tree under the C20+gamma model found using IQ-TREE, with full ultrafast bootstrap support values, see supplementary figs. S12–S24, Supplementary Material online.