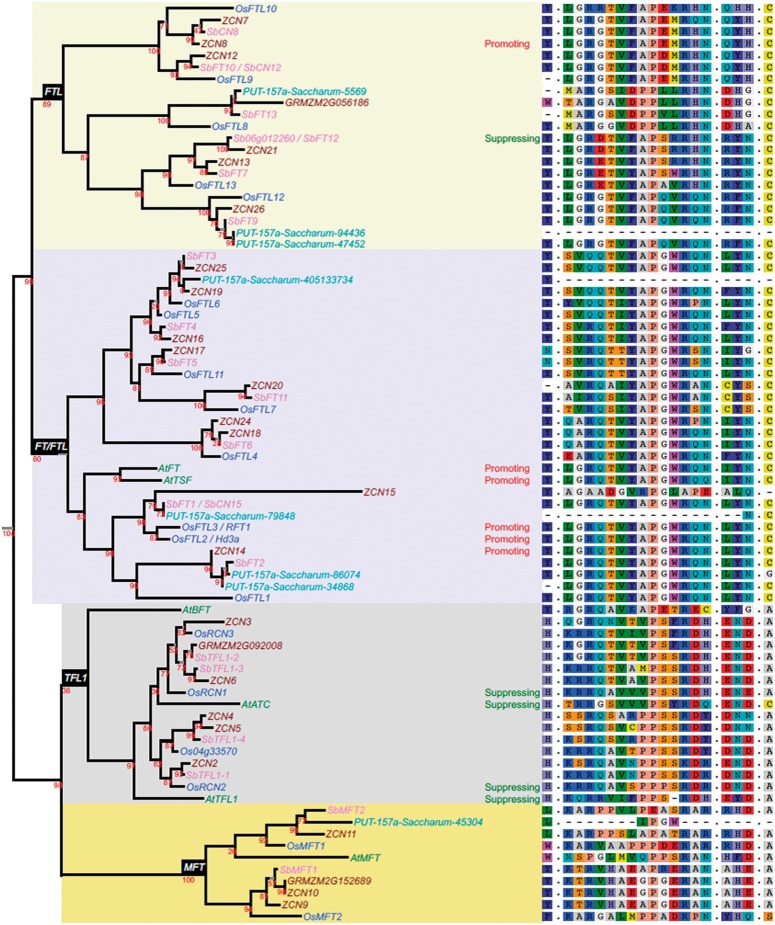
Fig. 3.
Homologs of Sb06g012260. Six homologs were found in Arabidopsis (prefixed At, including FT; Kardailsky et al. 1999), 19 in rice (Os, including Hd3a; Kojima et al. 2002), 19 in sorghum (Sb), 26 in maize (GRM or AC) and 8 in sugarcane (PUT-157a-Saccharum_officinarum), identified by BLAST from http://www.plantgdb.org/prj/ESTCluster/progress.php>PlantGDB, last accessed June 22, 2016. A phylogenetic tree of inferred homologous protein sequences was made at http://www.phylogeny.fr/, last accessed June 22, 2016, using MUSCLE for alignment and maximum-likelihood (PHYML) to determine the tree. Numbers on internal nodes indicate support values with 1000 bootstrap samples. Four distinct sub-families including FTL-like, FT/FTL-like, TFL1-like and MFT-like proteins, are highlighted on the tree. The impact on flowering (“promoting” or “suppressing”) are shown for several well studied PEBP genes (reviewed in Klintenäs et al. 2012) and for Sb06g012260/SbFT12. The panel on the right along the tree shows multiple alignments based on the Arabidopsis FT protein at amino acid position 85 at exon 2, positions 128–141 (P-loop domain), positions 150–152 and position 164 that were shown to have critical regulatory roles. For full alignments across the entire length of these proteins, see supplementary figure S4, Supplementary Material online.