Fig. 8.
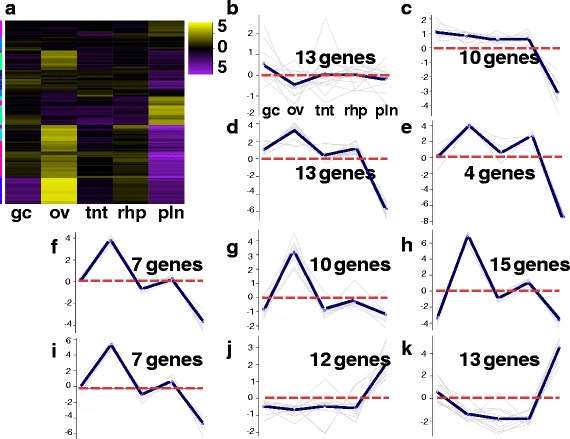
a-k Sex Heatmap for A. alata. Hierarchical clustering (EdgeR) and corresponding ten subcluster profiles for the 104 genes implicated in sex and early development differentially expressed across A. alata medusa (gastric cirri, ovaries, tentacle (and pedalium base), rhopalium) and planulae samples. Intensity of color indicates expression levels for each of the ten hierarchical clusters (vertical access). Bright yellow patches correspond to the highest peaks for each k-mean subcluster profile. K-mean profiles (b-k) match the order of column names in a, representing the mean expression of gene clusters highly abundant in each sample (centroid demarcated by the solid line; zero indicated by the horizontal dashed red line). Two yellow gene clusters in the gastric cirri column correspond to peaks in plot b and c; six bright yellow clusters in the ovaries column corresponds to profiles d-i; no major abundant gene clusters were detected in the tentacle column; four less intense clusters in the rhopalium column correspond to peaks in plots b, d, e, and h; two bright clusters in the planulae column correspond to peaks in the subcluster profiles j and k. The vertical colored bar on the left of the heatmap (a) indicates distinct patterns corresponding to the ten subcluster profiles (sc = subcluster number), for which the number of genes each comprises is indicated. Abbreviations: gc = gastric cirri, ov = ovaries, tnt = tentacle (and pedalium base), rhp = rhopalium, and pln = planulae. Gene annotations by subcluster provided in Additional file 9
