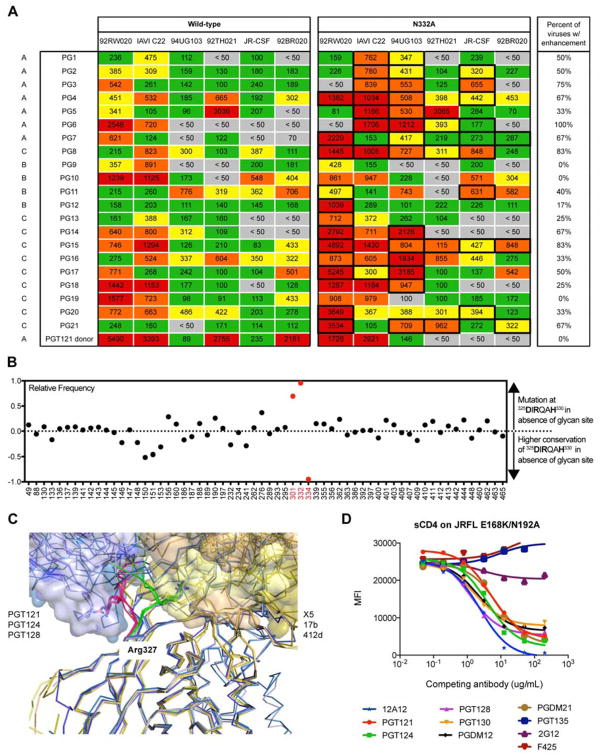
Figure 6. V3-glycan antibodies allosterically inhibit CD4 binding.
(A) Sera from 21 donors of the IAVI protocol G cohort were tested for neutralization on a 6-virus cross-clade panel of pseudoviruses with and without the glycan at the N332 position. Listed values are serum neutralization IC50. Neutralization titers that showed higher potency in the absence of the N332 glycan compared to the wild-type virus are highlighted with a bold box, and the percentages of viruses that resulted in enhanced neutralization potency in the absence of the N332 glycan site are listed. Serum from the PGT121 donor (N332 glycan dependent) was included as a control. (B) Among 42,715 HIV Env sequences in the Los Alamos database, 23,158 (54%) have the “GDIRQAH” sequence, while 19,557 (46%) deviate at D325, R327, or H330 residues. Using 46% as a baseline measurement of mutation at these residues by chance, every glycan site on Env was then evaluated for greater mutation (> 0) or greater conservation (< 0) of these residues in the absence of individual glycan sites. (C) Key contacts to residues 324GDIRQAH330 on gp120 by GDIR-bnAbs and CD4i antibodies. Overlaid liganded structures of PGT122, PGT124 and PGT128 (shades of blue) with key contacts to the 324GDIRQAH330 residues on gp120 highlighted in red. These structures are overlaid with liganded structures of CD4i antibodies 17b, X5, and 412d (shades of yellow) with key contacts to the 324GDIRQAH330 residues on gp120 highlighted in green. Arg327 on gp120 for all structures is shown as sticks. All structures are aligned on gp120. (D) High-mannose patch antibodies were tested for competition with sCD4 on isolate JR-FL E168K/N192A Env displayed on the surface of 293T cells. The CD4 binding site antibody 12A12 was included as a positive control and the V3-specific antibody F425 was included as a negative control.