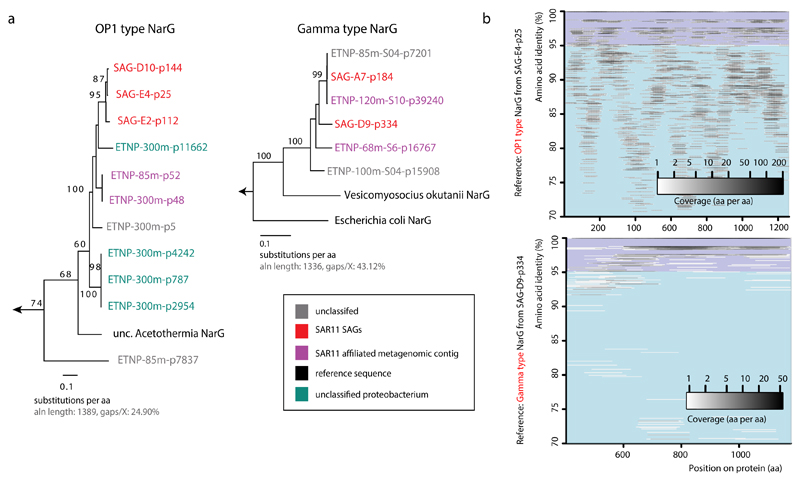
Extended Data Figure 7. Diversity of OP1 and Gamma-type narG amino acid sequences in the ETNP OMZ metagenome.
a, Phylogenies showing all full-length narG sequences recovered in the ETNP OMZ metagenomes (85, 100, 125, 300 m), as well as those from the SAR11 SAGs and corresponding narG reference sequences, with the left tree showing OP1-type variants and the right tree showing Gamma-type variants. NarG sequences are color-coded based on the taxonomic classification of adjacent genes in the same metagenomic scaffolds, as show in Supplementary Table 6. b, Recruitment of metagenomic reads (predicted open reading frames) from the OMZ 300 m sample, against OP1 (left) or Gamma (right) type narG sequences from the SAR11 SAGs. The metagenomic reads used for recruitment were identified as “narG” using the ROCker pipeline, and their identity further confirmed by phylogenetic placement within the narG clade on a reference DMSO superfamily protein tree, in order to minimize non-specific recruitments in conserved protein regions. Notice that based on this analysis, the OP1 type narG variants are highly diverse in the OMZ metagenome.