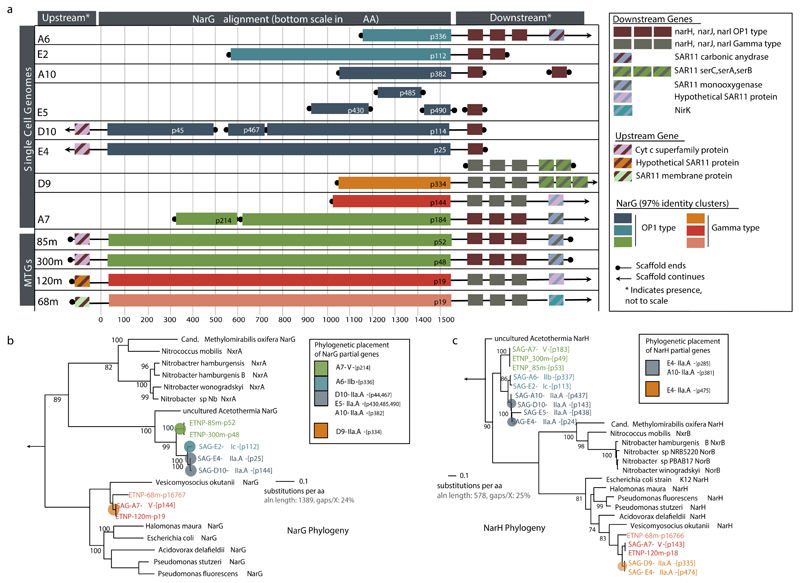
Extended Date Figure 3. nar genes encoded by SAR11 populations of OMZs.
a, nar operon and adjacent genes identified in SAR11 single amplified genomes (SAGs) from the ETNP OMZ, and in assemblies from the 85 m and 300 m ETNP OMZ metagenomes. narG sequences with at least 97% amino acid similarity are represented with the same color. b,c, Representative maximum likelihood phylogeny to show sequence variation among full or near full-length narG (b) and narH (c) amino acid sequences identified in the SAGs. A subset of cytoplasm-oriented nitrate reductases and nitrate oxidoreductases from publicly available genomes is also included. A comprehensive phylogeny showing the placement of SAR11 nar sequences relative to enzymes (n=392) of the DMSO family is in Fig. 2a. Colored pies represent the placement of shorter narG/narH gene fragments identified in the SAGs. Bootstrap values over 50 are shown. Outgroups (arrows) are Escherichia coli dmsA (b) and dmsB (c). Note: the Gamma-type nar-containing contig recovered in E4 (Fig. 2a) contains narHJI, but not narG; E4 Gamma-type is therefore not represented in Fig. 3b. All genes co-localized in the nar-containing contigs are listed in Supplementary Table 5. The p-numbers are gene IDs given by the gene prediction software, consistent with those in Supplementary Table 5.