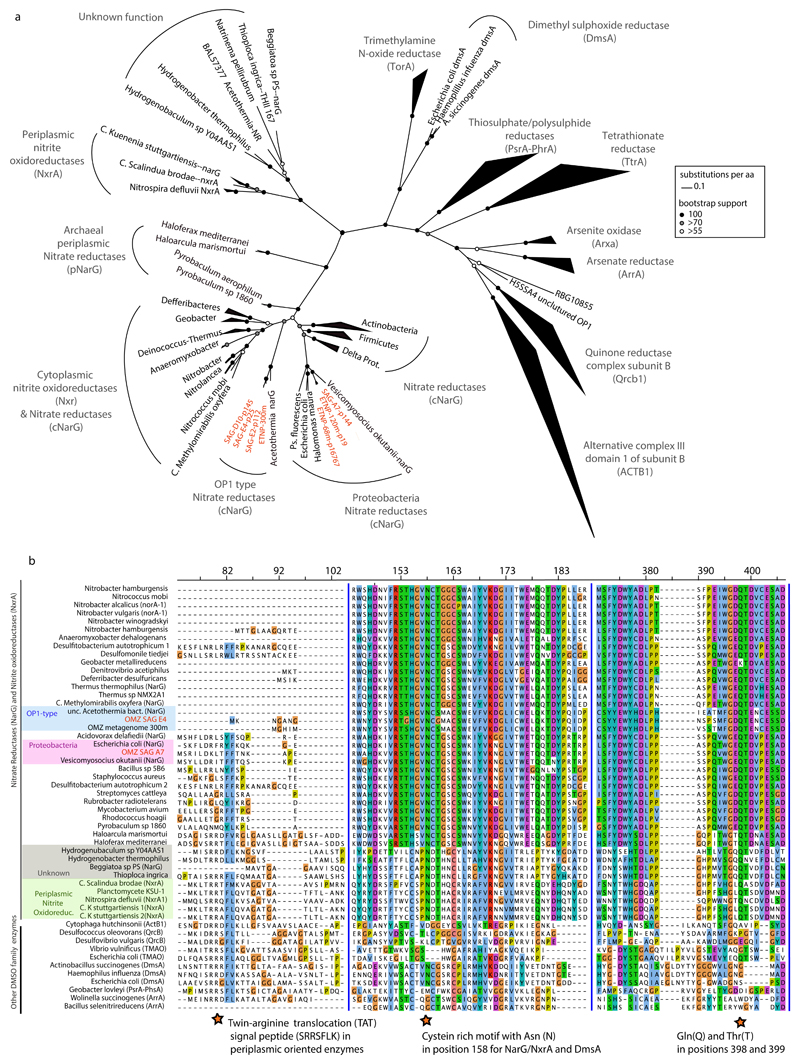
Extended Data Figure 4. Identified NarG in SAR11 SAGs are members of the DMSO superfamily of oxidoreductases.
a, Phylogenetic reconstruction of NarG and DMSO enzymes. The tree shown in Figure 2 is presented here but has been expanded to include diverse DMSO oxidoreductases for direct comparison with the NarG/NxrA enzymes. Notice that both OP1 (green, blue, grey) and Gamma-type (red, orange) variants cluster within the cytoplasmically oriented nitrate reductases and nitrite oxidoreductases. 697 NarG/NxrA proteins were identified from UniRef 63, and from those 321 full length sequences were selected to represent all the diverse clades. An additional 71 non-NarG/NxrA proteins, representative of the diverse enzymes of the DMSO superfamily were also included in the collection. The full-length amino acid sequences were aligned with Clustal Omega 61 and the phylogenetic tree was constructed by maximum likelihood and 1000 bootstraps using RAxML 60. The alignment length was 1803 columns, out of which 31.2% were gaps or undetermined. Partial NarG sequences identified in the SAGs were placed on the tree using the epa algorithm from RAxML 66. The same collection of proteins was used to train the Rocker models and quantify the narG metagenomic fragments, and can be found in the enve-omics website (http://enve-omics.ce.gatech.edu/rocker/models). b, Alignment of NarG sequences from OMZ SAR11 with representative sequences from the DMSO superfamily of oxidoreductases. The protein motifs in the second and third panels are present in all functional nitrate reductases (NarG) and nitrite oxidoreductases (NxrA) but not in closely related enzymes of the DMSO superfamily. The first panel shows the presence/absence of the TAT signal peptide (SRRSFLK), whose presence typically denotes a protein excreted to the outer membrane 40,41. SAR11 NarG is instead oriented toward the cytoplasm (lack of TAT). The second panel shows the cysteine-rich motif typically found in the N-terminus of the type-II DMSO superfamily oxidoreductases 76 and believed to enable the formation of a [4Fe–4S] cluster in these proteins 77. The Asn (N) in position 158 of the alignment is typically found in catalytic subunits of nitrite reductases and DMSO oxidoreductases (DmsA) but not in other DMSO family enzymes. The third panel shows the Gln(Q) and Thr(T) in positions 398 and 399 within the putative substrate entry channel of the protein, which differentiate the Nar proteins from all other oxidoreductases of the DMSO family 40.