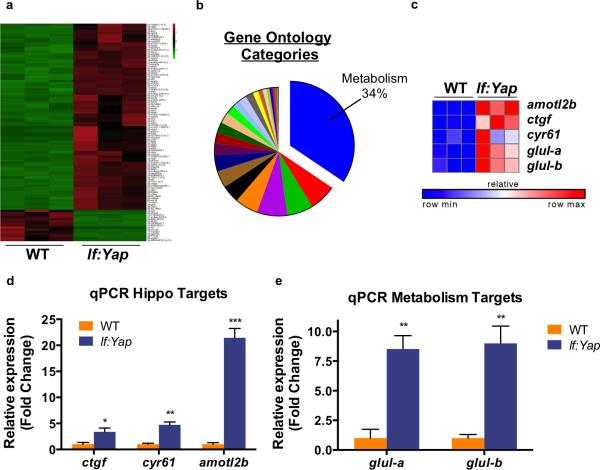
Figure 2. Yap alters expression of metabolism-related genes and enhances glul expression.
(a) Heatmap of statistically significant differential gene expression between dissected WT and lf:Yap livers. Green indicates decreased expression, while red demonstrates increased expression.
(b) Pie chart of Gene ontology (GO) categories upregulated by Yap. Biological processes related to metabolism are represented in blue.
(c) Heatmap analysis of RNAseq data illustrates the induction of individual Yap target genes (ctfg, amotL2, cyr61) and nitrogen metabolism genes glula and glulb.
(d) qPCR validation of Yap target gene expression in dissected WT and lf:Yap transgenic livers. n=4 WT and lf:Yap adult livers (with respect to ctgf expression). n=3 WT and lf:Yap adult livers (with respect to cyr61 and amotl2 expression); see Supplementary Table 4. *p<0.05, **p<0.01, ***p<0.001, two-sided Student's t-test, values represent the mean±SEM.
(e) qPCR validation of Glul induction in dissected WT and lf:Yap transgenic livers. n=3 WT and lf:Yap adult livers; see Supplementary Table 4. **p<0.01, two-sided Student's t-test, values represent the mean±SEM.