ABSTRACT
In this study, we investigate the effect of miR-34a expression and biological characteristics of breast cancer stem cells (BCSCs). The mammospheres were formed from murine breast cancer cell line 4T1 and regarded as murine BCSCs. Identification of stemness molecules and cloning experiments validate the biological characteristics of BCSCs we have established. We showed that miR-34a, as a tumor suppressor, could separately reduce the stemness of BCSCs and activate the cytotoxic susceptibility of BCSCs to natural killer (NK) cells in vitro via down regulating the expression of Notch1 signaling molecules. Moreover, miR-34a could completely restrain established mice breast tumor xenografts in vivo in the NOD/SCID mice that have functional NK cells at a normal level, whereas it was less effective in NOD/SCID/ CD122/IL-2Rβ mice that do not have functional NK cells. We conclude that miR-34a is a crucial, dual tumor suppressor and BCSCs-targeting immunotherapeutic agent and has shown efficacy in the treatment of murine breast cancer. The results also suggest that impaired NK cells could contribute to the resistance to therapies.
KEYWORDS: Cytotoxic susceptibility, ligand, miR-34a, Notch1
Introduction
Cancer stem cells (CSCs) possess a unique character to initiate malignant growth, which provided a vital insight to recognize origin and essence of tumors. MicroRNAs (miRNAs), a class of conservative, short and endogenous non-coding RNAs of approximately 19–26 nt, play critical roles in tumorigenesis via down-regulating target genes.1 The effective regulatory of miRNAs is of intriguing interest on CSCs research. Breast cancer represents the most common cause of cancer related lethality in women throughout the world with very low five-year survival rates, even after clinical therapy.2 Through the gene expression profile analysis of breast CSCs (BCSCs) and non BCSCs miRNAs, we learned that the expression of miR-125, Let-7, miR-21, miR-155 and miR-34a were decreased obviously in BCSCs. Undoubtedly, the study of miRNAs on BCSCs will provide the probability of radical curing breast cancers through the targeted killing of CSCs.
miR-34a is highly expressed in normal tissues and commonly repressed in carcinoma such as prostate cancer,3 colon cancer,4 breast cancer,5 ovarian cancer and non-small cell lung cancer.6,7 Moreover, previous studies demonstrated that ectopic expression of miR-34a suppressed cell proliferation,8 migration and invasion in various cancer cells,3,9 which could also contribute to drug resistance in breast cancer by targeting a variety of oncogenes.10 However, the role and mechanism of miR-34a in the regulation of BCSCs is far from being completely elucidated at present. Notch signaling is an evolutionarily conserved signal pathway. The four members of Notch receptors have been identified to date in vertebrates, including Notch1-411. Among them, Notch1 is not only involved in normal cells differentiation but also related to tumors proliferation and survival. According to TargetScan, Notch1 is a direct target of miR-34a and previous research showed that miR-34a could combine with the 3′-UTR region of Notch1 mRNA sequence, and then inhibit the protein expression of Notch1 and downstream molecules.12,13 Another report has alleged that aberrant Notch1 signaling could facilitate the proliferation and survival of BCSCs.14 In this study, we intend to explore whether miR-34a can reduce the stemness of BCSCs by targeting and further inhibiting Notch1 and its downstream molecules.
On the other hand, NKG2D is activating receptor and is critical for NK cells to kill tumor cells. In mice, NKG2D is mainly to identify and combine retinoic acid early transcription factor 1 (Rae1), histocompatibility 60 antigen (H60a) and Ulbp1 binding protein1 (MULT1).15 Wang et al invented that human tumor-infiltrating NK cells failed to limit metastasis and were not associated with improved therapeutic outcome of BCSCs-rich breast cancer.16 They found primary BCSCs could escape NK cell killing effect of biological characteristics, MICA and MICB, two human ligands for the stimulatory NK cell receptor NKG2D, expression down-regulation was the main reason. In this study, we intends to investigate whether miR-34a can upregulate the expression of murine NKG2D ligands (NKG2DLs, such as Rae1, H60a and MULT1 in BCSCs and then indirectly boost NK cell killing effect to BCSCs, a new strategy may make it possible to eliminate the remaining BCSCs and cure cancer thoroughly, and we hope to be the result of killing two birds with one stone.
Materials and methods
Mice, cell culture and murine primary NK cells
Female NOD/SCID mice that were 4 weeks old were purchased from Beijing Animal Center. Animal experiments were done according to the Institute Research Ethics Committee at the Nankai University. The metastatic murine breast cancer cell line, 4T1, was purchased from ATCC. 4T1 cells were cultured in RPMI-1640 supplemented with 10% fetal bovine serum (FBS, BI, Israel, South America). 4T1 cells were grown to 90% confluence, trypsinized, and plated at a density of 5,000 cells/ml in serum-free Dulbecco's modified Eagle's medium (DMEM)/Ham's F12 medium (1:1) (Hycolone, Los Angeles, USA) supplemented with 1× B27 (GIBCO, Grand Island, NY, USA), 20 ng/ml epidermal growth factor (EGF, Dakewei, Beijing, China), 10 ng/ml basic fibroblast growth factor (bFGF, Dakewei, Beijing, China), 5μg/ml insulin (Beyotime Biotechonlogy, Jiangsu, China), 0.4% bovine serum albumin (BSA, Sigma, Shanghai, China), 100 units/ml penicillin and 100 mg/ml streptomycin in 25 cm2 non-adherent flask.17,18 Murine primary NK cells were isolated from splenocytes of six-to-eight week old female BALB/c mice by EasySep Selection Mouse NK Cell Enrichment Cocktail (Stem Cell Technologies, Grenoble, France). Then, murine primary NK cells were grown in RPMI-1640 supplemented with heat-inactivated 10% FBS, 200U/ml recombinant mouse IL-2 (rIL-2, Dakewei, Beijing, China), 10µM-mercaptoethanol, 100 units/ml penicillin and 100 mg/ml streptomycin.
Prediction of microRNA-binding sites
The predicted microRNA binding sites were downloaded from TargetScan 5.1 Mouse (http://www.targetscan.org/mmu_61/) and microRNA.org (http://www.microrna.org/microrna).
RT-PCR and quantitative real time PCR analysis (qPCR) analysis
1 μg of RNA was reverse transcribed with the EasyScript First-Strand cDNA Synthesis SuperMix Kit (TransGen, Beijing, China). Then 1 μl cDNA was used for RT-PCR with 2×EasyTaq PCR SuperMix Kit (TransGen, Beijing, China), qPCR was performed with the QuantiFast SYBR Green PCR kit (Qiagen, Valencia, CA, USA). Mouse β-actin was performed on each experimental sample as an endogenous control. Specific stem-loop primers were used for reverse transcription reaction of miR-34a. The RNU6 was an internal control for normalizing the expression of miR-34a. All primer used for experiment was summarized in Table 1 (S1). Statistical results were averaged from three independent experiments performed in triplicate.
Transient transfection with synthetic oligonucleotides (oligos)
MiR-34a mimic, inhibitor and negative control oligonucleotides were purchased from RiboBio (Guangzhou, China). The transfection of mouse BCSCs with 50 nM mimic or negative control were performed with LipoJet™ Reagent (SignaGen Laboratories, JiangSu, China) according to the manufacturer's protocol. The transfected cells were harvested for study after culturing for 48 h.
Cell proliferation analysis by Cell Counting Kit-8 (CCK8)
Briefly, BCSCs were seeded into 96-well plates at 2,000 cells per well and transfected with miR-34a, miR-NC oligos, and anti-34a or anti-NC oligos. The effects of miR-34a on cell proliferation were detected 48h after seeding using CCK8 (Beyotime Biotechonlogy, Jiangsu, China). Each assay was performed in three replicates on three independent experiments.
Clonal and clonogenic assays
For holoclone assays, we plated the secondary generation of BCSCs at a clonal density (i.e., 1,000 cells/well) in a 6-well dish and treated with miR-34a mimic or negative control oligos. The number of holoclones was counted 5–7 days later, and the percentage of cells that established a holoclone was presented as cloning efficiency. For clonogenic assays, firstly, we added 0.5% agar (diluted 1% agar with serum-free DMEM/F12 medium) in 6-well ultra-low attachment plates as the bottom layer and incubated at room temperature for 30min. Then we mixed the transfected BCSCs (1,000 cells/well) with the 0.375% agar (diluted 1% agar with serum-free DMEM/F12 medium) and put them upon the bottom layer to form middle layer. In the end, serum-free DMEM/F12 medium, as the outermost layer, was added to provide nutrition and enumerated colonies 2–3 weeks after plating. For all above experiments, we run a minimum of triplicate wells for each condition.
Flow cytometry (FACS)
BCSCs were transfected with olgios for 48h, then cells were collected and resuspended at 1×106 in 100 μl FACS buffer (PBS supplemented with 2% FBS), and incubate with primary rabbit polyclonal antibody to Ki-67 (Cell Signaling Technology, Danvers, USA) for 1h at room temperature. The cells were washed with PBS and centrifuged at 800rpm for 5min and then incubated with TRITC-conjugated secondary antibody for 30min. For apoptosis assays, cells were incubated with Annexin V-FITC and PI (BD Bioscience, New Jersey, USA) for 15 min in room temperature according to the manufacturer's instructions; For purification and elimination experiments, NK cells were incubated with anti-mouse NK1.1 FITC and anti-mouse CD3 PE antibody (eBioscience, SanDiego, CA) for 15min. The cells were measured using FACSCalibur and analyzed with FlowJo software.
Western blot analysis
Briefly, 30 μg of total protein was separated on 10% or 6% sodium dodecyl sulfate-polyacrylamide gels (SDS-PAGE) and electrophoretically transferred to a polyvinylidene difluoride (PVDF) membrane and incubated overnight at 4°C with rabbit anti-Nanog, anti-Oct4 (Abcam, Massachusetts, USA), anti-H60 (R&D Systems, Minneapolis, MN, USA), anti-Sox2 or goat anti-Notch1 (Santa Cruz Biotechnology, CA, USA). Then the membranes were incubated with 1:3000 dilution of HRP-conjugated goat anti-rabbit (Santa Cruz Biotechnology, CA, USA) or rabbit anti-goat (Bioworld, Shanghai, China) secondary antibody for 2 h at room temperature. The polypeptides were revealed using ECL reagent (Millipore, Bedford, MA,USA) and evaluated using the Chemilucent Plus Western Enhancing Kit. An anti-GAPDH (Santa Cruz Biotechnology, CA, USA) antibody was used as a protein loading control.
Adhesion assay
MicroRNA transfected BCSCs were seeded at a density of 1×104 per well in a 6-well plate to form a monolayer. Primary murine NK cells from splenocytes were added at a density of 5×104 per well and cocultured overnight. After removal of nonadherent cells by PBS washing, the adherent cells were counted under a microscope in 10 high-power fields. Photos were taken at ×400 magnification.
CD107a and Perforin 1 secretion assay
BCSCs were collected after 48 h transfection, and then co-cultured with primary murine NK cells (5×104 cells/well) at 1:5 ratio in 6-well plates, the cell culture supernatants of different groups were harvested at hours 8, 16, 24 and 32 and analyzed using mouse ELISA Kits (Elabscience, Wuhan, China) for LAMP-1 (Lysosomal Associated Membrance Protein 1, CD107a) and Perforin 1 (PRF1) according to the manufacturer's instructions. All samples were assayed in duplicate.
Cytokine secreting assay
The 4T1 breast cancer stem cell-conditioned medium was collected 48h after the transfection of miR-34a mimic or negative control. Cytokines secreted by 4T1 BCSCs were detected by the Mouse Cytokine Array Panel A (R&D, Minneapolis, MN, USA) based on the Proteome Profiler Array Protocol. Cytokine array data on the ChemiScope3600 Mini chemiluminescence imaging systems (Clinx Science Instruments Co., Ltd. Shanghai, China) was analyzed by the array image file using ImageJ software (http://rsbweb.nih.gov/ij/).
Tumor model
The effect of miR-34a with/without NK cells on breast cancer was determined in a murine breast carcinoma model. Briefly, rat anti-mouse CD122/IL-2Rβ (200 mg/mouse, GenWay Biotech, Inc. Beijing, China) was injected intraperitoneally (i.p) 1 day before tumor cell injection to eliminate residual murine NK cells as “without NK cells group.” Then, we acutely purified BCSC cells from 4T1 cells and transfected with miR-34a or miR-NC oligos (50 nM) 48 h later, 106 cells each were subcutaneous injected (s.c) in female NOD/SCID mice (with/without NK cells groups). On d7, 13, 20 and 25, miR-34a or miR-NC oligos mixed with LipoJet™ Reagent were injected intratumorally. We also purified BCSCS from xenograft tumors and transfected with miR-NC or miR-34a oligos. 24 h later, 106 cells each were injected s.c in female NOD/SCID mice which were pretreated like first generation. On day 28, the tumor of each mouse was harvested for measurement and immunohistochemical analyses.
Immunohistochemical (IHC) analysis
Paraffin-embedded and formalin-fixed tumor samples were cut into 5 μm sections, and then processed for incubation with anti-Notch1 Ab, immunoactivity was detected with diaminobenzidine (DAB).
Statistics
Results are presented in the form of “mean ± SE” and were analyzed with SigmaStat10.0 software (SPSS). The differences between 2 groups were assessed by the Student t test. The differences among 3 or more groups were evaluated by a one-way ANOVA followed by the Dunnett test. P < 0.05 was considered statistically significant.
Results
Identification and cultivation of mice breast cancer stem cells (BCSCs)
Murine breast cancer cell line 4T1 cells were seeded on culture flask and cultured in serum free medium, 24 h later, part of cells went into a state of apoptosis for failing to adapt to the serum free medium environment, while the rest of living suspension cells began proliferation, and the mammospheres formation could be observed obviously with microscope by culturing for 3 days, each mammosphere consisted about 50 cells, and the mammospheres became more regular, the size became larger, furthermore the number reached a hundred or more in each mammosphere after one week of culture (Fig. 1A).
Figure 1.
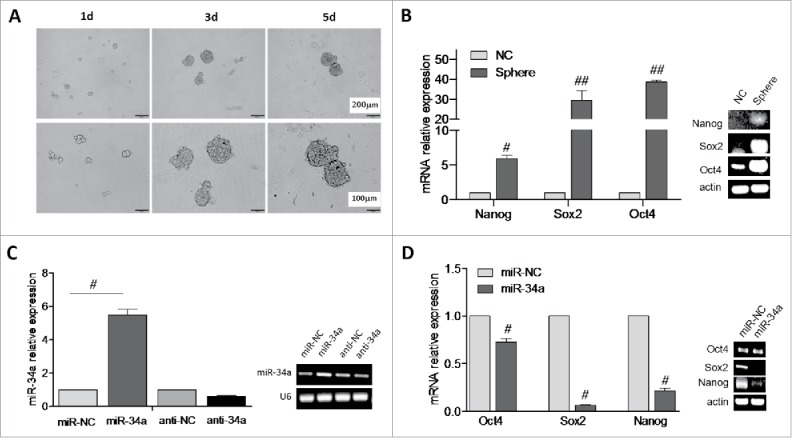
Over expression of miR-34a reduce the stemness of BCSCs. (A) Image of BCSCs mammospheres formation. (B) The relative expression of Nanog, Sox2 and Oct4 in 4T1 spheres and 4T1 cells (NC) was analyzed by RT-PCR and qPCR. (C) miR-34a expression level in spheres was determined by RT-PCR and qPCR. (D) The relative expression of Nanog, Sox2 and Oct4 in spheres transfected with miR-34a or miR-NC was evaluated by RT-PCR and qPCR. #P < 0.05, ##P < 0.01.
In order to identify the stemness of mammospheres, we assessed Sox2, Nanog and Oct4 mRNA expression in both mammospheres and 4T1 cells by RT-PCR and qPCR. The expression levels of stemness-related genes Sox2 and Oct4 were highly skyrocketed in mammospheres (P < 0.01), Nanog was also enhanced in mammospheres (P < 0.05) (Fig. 1B). Ulteriorly, soft agar assay revealed that the cloning efficiency of mammospheres was higher than 4T1 cells (S2).
The inhibitory effect of miR-34a on mice BCSCs
MiR-34a has been reported to be a tumor-suppressor in the inhibiting tumorigenic subpopulations of CD44+ prostate CSCs.3 To better understand whether miR-34a had the potential biological functions of miR-34a in BCSCs, the BCSCs were transfected with synthetic mature miR-34a, miR-NC oligos, and anti-34a or anti-NC oligos for 48 h. The mRNA level of miR-34a was assessed by RT-PCR and qPCR. As expected, miR-34a mimic transfected BCSCs showed miR-34a levels higher than cells with miR-NC (P < 0.05). In contrast, miR-34a inhibitor transfected BCSCs showed reduced endogenous miR-34a (P < 0.05) (Fig. 1C). Then we identified the expression of Sox2, Nanog and Oct4 mRNA markedly deceased in miR-34a mimic transfected BCSCs (P < 0.05) (Fig. 1D). Thus it suggested miR-34a over expression in BCSCs could reduce their stemness leading to CSCs depletion and senescence.
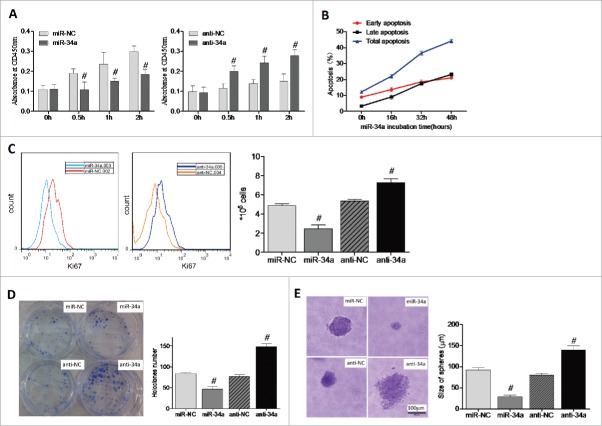
To further investigate the effects of miR-34a on BCSCs properties, we verified the effect of miR-34a on the proliferation and apoptosis of BCSCs. Results showed that miR-34a overexpression could significantly suppress BCSCs proliferation after 0.5 h, 1 h, 2 h, respectively. We measured the BCSCs absorbance at OD450nm which reflected the proliferation rate of cells. The quantified data shown that miR-34a inhibited the proliferation of BCSCs compared with miR-NC, while anti-34a had the opposite effect compared with anti-NC (Fig. 2A). miR-34a over-expression in cells caused enhanced apoptosis at 16 h, 32 h and 48 h wherein by FACS we detected early and late apoptosis. The miR-34a overexpression led to same tendency in early and late apoptosis at three time points, and with the highest rate of total apoptosis for 48h incubation with miR-34a (Fig. 2B, S3). Similar results were obtained from FACS that miR-34a could obviously suppress BCSCs proliferation (P < 0.05) (Fig. 2C). Then we carried out the holoclone and clonogenic assays to detect the self-renewal and mammospheres formation ability with the secondary generation of BCSCs. The results revealed that miR-34a over expression inhibited holoclone formation of BCSCs, while anti-34a played the opposite effects on BCSCs (P < 0.05) (Fig. 2D); Moreover, soft agar assay analysis showed that the clonogenic capacity of BCSCs which transfected with miR-NC were higher compared with miR-34a mimic transfected BCSCs (Fig. 2E). The above experimental results provided evidence that restoration of miR-34a expression in BCSC cells inhibits proliferation, clonal and clonogenic self-renewal, it embodied miR-34a was a negative regulator of the tumorigenic properties of BCSCs.
Figure 2.
Overexpression of miR-34a lower the self-renewal and mammospheres formation of BCSCs. (A) The effect of miR-34a or anti-34a on proliferation of BCSCs was detected by CCK8. (B) The effect of miR-34a on the early and late apoptosis of BCSCs was quantified by FACS. (C) Ki-67 expression of BCSCs transfected with miR-34a or anti-34a was analyzed by FACS. (D, E) Clonal and Clonogenic assays of BCSCs transfected with miR-34a or anti-34a were performed, respectively. (n = 3). #P < 0.05.
miR-34a inhibits the expression of Sox2, Oct4 and Nanog by targeting Notch1 pathway
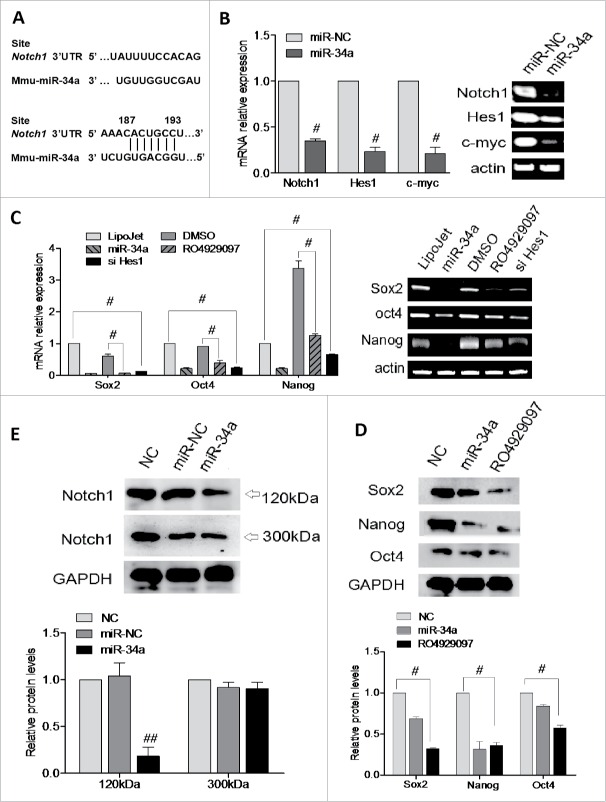
To further investigate the molecular mechanism of miR-34a, a search for potential miRNAs that target the 3′UTR of Notch1 was based on the miRBase and TargetScan software prediction. We obtained that Notch1 had a binding site for miR-34a in its 3′UTR region (Fig. 3A). The result of RT-PCR and qPCR revealed that transfection of miR-34a reduced the RNA level of Notch1 and its downstream molecules Hes1 and c-myc (Fig. 3B). Which could demonstrate miR-34a negatively modulated Notch1 pathway. Next, we found Sox2, Oct4 and Nanog mRNA and protein expression was lower in the RO4929097 (a Notch1 inhibitor) or siHes1 treatment (Fig. 3C and 3D), and which suggested Nocht1 or Hes1 played a role in down-regulating the expression of Sox2, Oct4 and Nanog. Here, we speculated that miR-34a reducing stem cell stemness in BCSCs might be associated with targeting Notch1 pathway. In addition, the expression of Notch1 (300 kDa) and Notch1 (120 kDa), known as ICN (the intracellular domain of Notch1) was elevated, and we found that the expressions of Notch1 (120 kDa, ICN) in miR-34a transfection group was significantly lower than those in miR-NC transfection group (P < 0.05), while Notch1 (300kDa) expression had no significant difference(Fig. 3E). RO4929097 was a γ-secretase inhibitor, γ-secretase was a key protein released by Notch intracellular domain (NICD), called ICN. To recap, we could definitely figure out that miR-34 suppressed the activation of Notch1 signaling pathway via mainly blocking the release of ICN.
Figure 3.
Effect of miR-34a on BCSCs is associated with a decrease in Notch1 and Hes1 expression. (A) Schematic of the Notch1 3′UTR indicates seed-matching sequences and miR-34a seed sequences. The black vertical bars indicate possible base pairing. (B, C) mRNA levels of Notch1, Hes1, c-myc, Sox2, Oct4 and Nanog under different conditions were analyzed by RT-PCR and qPCR (n = 4). Results are shown as the mean expression values normalized against miR-NC, LipoJet or DSMO. (D, E) Notch1(300KDa), Notch1(120KDa, ICN), Sox2, Oct4, Nanog and GAPDH in BCSCs transfected with miR-34a, anti-34a or inhibited by RO4929097 were detected by Western blot. (n = 3). #P<0.05.
miR-34a enhances the BCSCs cytotoxic susceptibility to NK cell
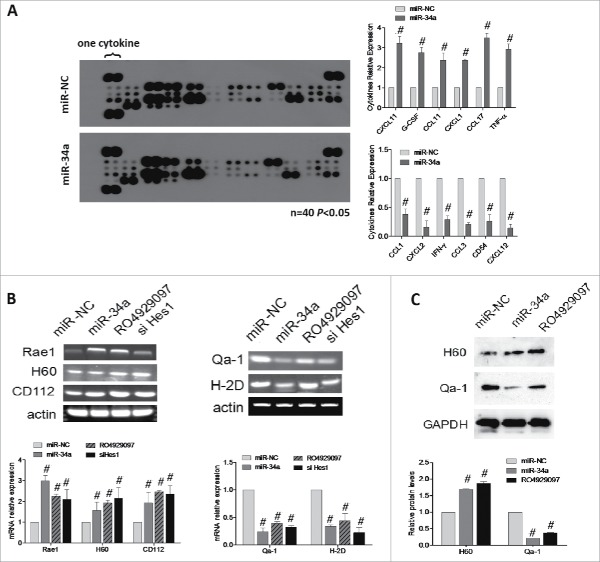
Researchers reported that in isolated bone marrow-derived macrophages (BMM) from Notch1+/− mice, the analysis of cytokines and chemokines in culture supernatants revealed that Notch1 deficiency results in decreased inflammatory cytokine expression.19 Here, we checked the change of cytokines along with BCSCs tranfected miR-34a. First, supernatants were collected from BCSCs transfected with either the miR-34a mimic or its miR-NC and used to detect cytokine levels by means of a mouse cytokine array panel (Fig. 4A). In fact, NK-related cytokines such as CXCL1 and CXCL11 were increased, when miR-34a was overexpressed in BCSCs (Fig. 4A right). These observations hinted that miR-34a might influence NK cells, since some of these cytokines related to the function of NK cells.
Figure 4.
Over expression of miR-34a enhances the cytotoxic susceptibility of BCSCs to NK cell. (A) Co-culture supernatants of NK cells and BCSCs transfected with miR-34a or miR-NC were collected and the cytokine levels were detected by the mouse cytokine array panel. (B) Rae1, H60, CD155, CD112, Qa-1, H-2K and H-2D mRNA levels were analyzed by RT-PCR and qPCR under different conditions. (C) Protein was extracted from spheres treated with miR-NC, miR-34a and RO4929097, then validated the expression of H60, Qa-1 by Western blot. #P < 0.05.
The activating receptors were expressed on the surface of NK cells, while their ligands were expressed on the surface of mice cancer cells or CSCs. We evaluated the cytotoxic susceptibility of BCSCs which were treated with miR-34a by detecting the RNA levels of H60, Rae1, CD155, CD112, H-2K, H-2D and Qa-1. RT-PCR and qPCR results showed that miR-34a could promote the ligands expression of activated receptors, such as H60, Rae1, CD155 and CD112, while inhibit the ligands expression of inhibitory receptors, H-2K, H-2D and Qa-1(Fig. 4B). So miR-34a-transfected BCSCs would be easier for NK cells to kill. Furthermore, we detected the expression change of above ligands of the CSCs treated with RO4929097, Notch1 inhibitor, and siHes1 in mRNA or protein level, we obtained the results of supporting Notch1 signaling molecules involved in miR-34a mediating the expression of NK cells ligands (Fig. 4B and C). These results indicated that miR-34a should make the NK cells overall robust.
NK cell adhension and cytotoxity increased in BCSC-miR-34a
The biofunction of NK cell was determined in vitro by analysis of cell-cell adhesion of murine primary NK cells to miR-34a-BCSCs. miR-34a-BCSCs and control cells were incubated with murine primary NK cells. miR-34a-BCSCs induced a 3.4-fold increase of adherent NK cells (Sparkles of light, arrows) compared with BCSC control (wild-type and NC), suggesting that miR-34a-BCSCs mediated firm adhesion of NK cells (Fig. 5A). Finally, we analyzed the secretion of CD107a (Fig. 5B) and PRF1 (Fig. 5C) by NK cells induced by miR-34a-BCSCs. The highest amount of CD107a and PRF1 was observed in the supernatant of NK cells co cultured with miR-34a-BCSCs (121 ± 33 pg/ml; 352 ± 63 pg/ml, respectively, mean±SD), in contrast to BCSC controls (97 ± 14 pg/ml, 207 ± 87 pg/ml, respectively). With miR-34a expression became low for anti-34a-BCSCs, the amount of CD107a and PRF1 showed the opposite tendency (90 ± 17 pg/ml; 211 ± 35 pg/ml, respectively, mean ± SD), in contrast to BCSC controls (112 ± 41 pg/ml, 452 ± 122 pg/ml, respectively). This finding associated miR-34a with the induction of cytotoxic tumor microenvironment.
Figure 5.
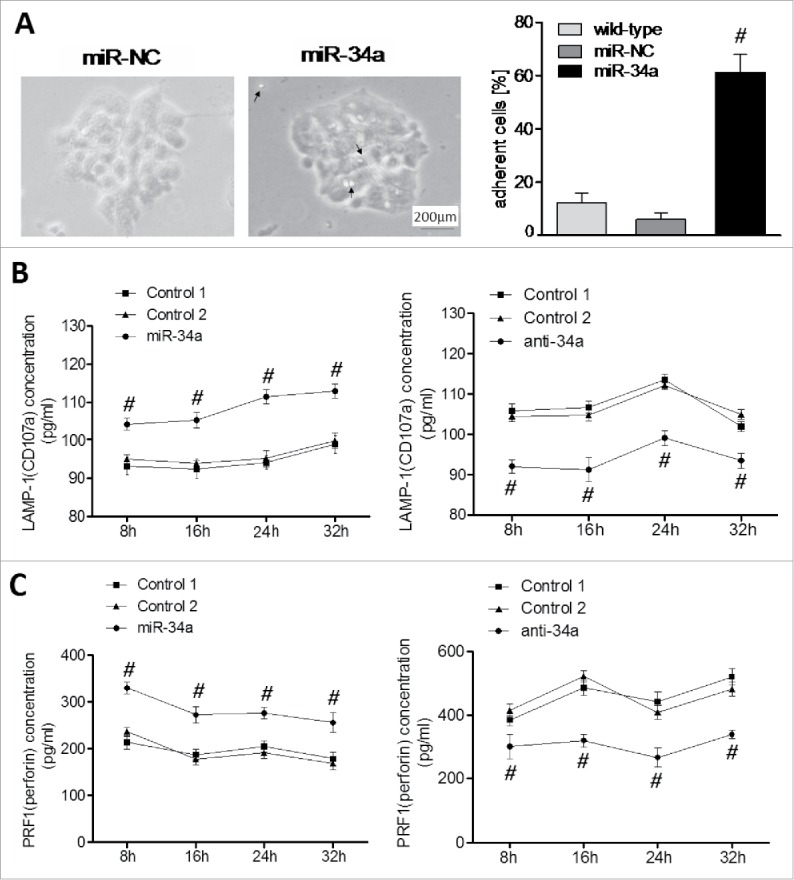
Adhesion and secretion of CD107a and Perforin of NK cells are induced by miR-34a. (A) Adhesion induced by miR-34a was determined by co-incubation of murine primary NK cells and miR-34a-BCSCs. Pictures of adhesion between miR-34a-BCSCs and NK cells after 12 h incubation were taken at x100. Sparkles and arrows highlighted NK cells adherent to miR-34a-BCSCs. (B) Secretion of CD107a and (C)Perforin from NK cells was determined by ELISA (n = 3). #P < 0.05.
Experiment in vivo
In order to investigate whether the anti-tumor effect of miR-34a was cooperated with the cytotoxicity of NK cells we performed mouse tumorigenicity assay. According to the presence of NK cells, we regarded NOD/SCID mice as control group and CD122/IL-2Rbeta treatment NOD/SCID mice as NK cell depletion group (90% NK depletion as determined by FACS, Fig. 6A right, S4). Our results showed that the formation rate of tumor was 100% as we expected. The tumor obviously grew slowest after injecting BCSCs transfected with miR-34a than miR-NC, the tumor weight decreased to 1.26±0.79 g (P < 0.05). Additionally, the tumor after injecting miR-34a with NK cell depletion visibly grew slower among the three groups, the tumor weight increased to 1.94 ± 0.13 g (P < 0.05) comparing with the NK cell presence group (Fig. 6B). miR-34a also inhibited the secondary transplantation of 4T1 BCSCS (Fig. 6B left). Paraffin section dying showed that Notch1 staining was primarily localized to the cell membrane and cytoplasm, suggesting that the protein was activated in tumor tissues. Notch1+ cells number of miR-34a treatment group had the lowest level than the control group and the NK cell absence group (Fig. 6C). At last, the RNA levels of the ligands H60, Rae1, CD155, CD112 of tumor tissues homogenate identified that miR-34a could promote the ligands expression of activated receptors, while inhibit the ligands expression of inhibitory receptors. miR-34a-transfected BCSCs would be more sensitive to NK cell cytotoxicity (P < 0.05, Fig. 6D). Together, these above results strongly suggested that the inhibiting effect of miR-34a on tumor growth emerged as the best in the presence of NK cells.
Figure 6.
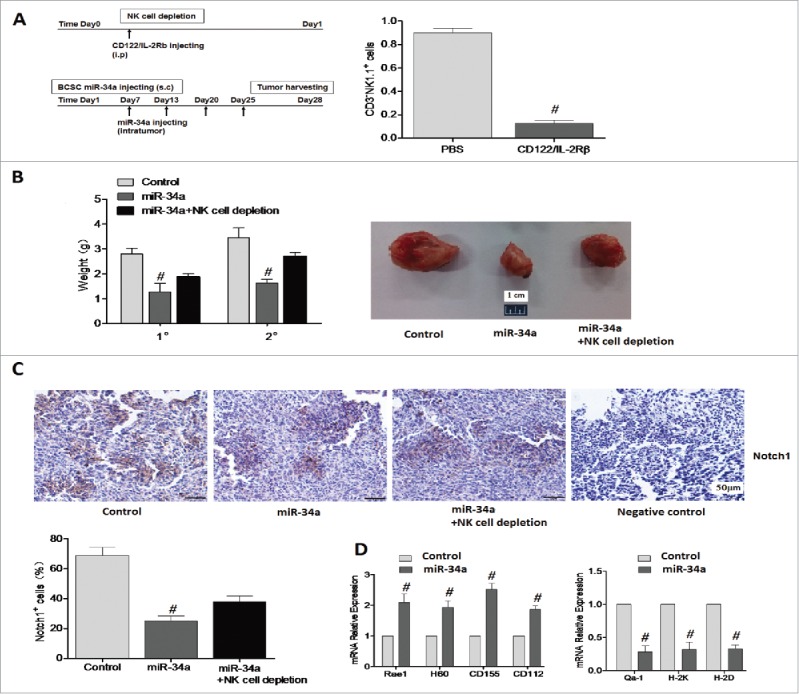
miR-34a cooperating with NK cells inhibit breast cancer cell growth in mice that were injected with miR-34a-BCSCs. (A) Schematic illustrating the methods of treating animal models in vivo. NK cells depletion in splenocytes was quantified by FACS. (B) Tumor weight in first (1°) and secondary (2°) transplantation of BCSCs was valuated (left), the tumors in first transplantation were showed (right). (C) Representative immunohistochemical analyses of Notch1 in the tumors dissected from control mice and miR-34a treatment mice. n = 6 per group. (D) Rae1, H60, CD155, CD112, Qa-1, H-2K and H-2D mRNA from tumor tissue homogenate of two groups was determined by qPCR (n = 4). #P < 0.05.
Discussion
CSCs, or tumor-initiating cells, are involved in tumor progression and metastasis.20,21 miRNAs regulate both normal stem cells and CSCs,22-24 and dysregulation of miRNAs has been implicated in tumorigenesis.25 In this study, we generated BCSCs from serum-free medium (SFM) and transfected with the synthetic mature miR-34a oligonucleotides (oligos). The results revealed that enforced expression of miR-34a inhibited BCSCs sphere establishment, holoclone formation, clonogenic capacity, proliferation and easier to be killed by NK cells. In contrast, miR-34a antagomirs promoted tumor development. Here, we confirmed these functional aspects of miR-34a, also presented how miR-34a eliminated BCSCs mainly in two directions: decreasing the stemness and improving the susceptibility to NK cell activity. Furthermore, we observed that miR-34a overexpression lead to dramatic downregulation of Notch1 and its target genes (Hes-1 and cmyc), which provide a functional link between a stemness impairment and an enhancement of susceptibility on BCSCs during this process.
The experts were now fully convinced that the miR-34a, either individually or in combination with other miRNA(s), inhibited cancer cells stemness and metastasis, but what cues did it follow? It was one thing we wanted to explore. Many researches revealed Notch1 signaling was always activated in haematopoietic stem cells,26 intestinal stem cells,27 muscle stem cells and bone mesenchymal stem cells.28,29 Beyond that, a report's analysis articulated that miR-34a inhibited cell proliferation by targeting Notch1 in renal cell carcinoma.30 Meanwhile, other reports suggested that P53 could combine with the promoter of miR-34a and activate its transcription. P53 mutate could inhibit miR-34a transcription activation and result in a lower expression of miR-34a.31 Otherwise numerous studies demonstrated there was a direct link between Notch1 and P53. These were good grounds for speculating that miR-34a might inhibit the stemness of BCSCs by mediating Notch1 pathway. Under this consideration, we searched the miR-34a binding site in the 3′UTR of Notch1 mRNA by TargetScan Mouse and microRNA.org software. The result was exciting. Then, we discovered that miR-34a over expression in BCSCs could silence its potential targeting molecules Notch1, Hes1 and c-myc, meanwhile the expression of Sox2, Nanog and Oct4 were decreased as well. Along with this change, miR-34a overexpression also exhibited lower clonal and clonogenic potentials and Ki-67+ BCSCs than miR-NC. These strongly suggested miR-34a inhibited stem cell proportion and cell proliferation by down-regulating Notch1 signaling in BCSCs. Other studies defined that miR-34a was down-regulated in several prostate cancer stem/progenitor cell populations, miR-34a over-expression could induce G1 phase cell-cycle arrest accompanied by cell senescence.32 These displayed that miR-34a as a certain key negative regulator of CSCs has been the growing interest of scientists.
Studying in another way, we investigated the change of cytokines in Notch1 dysregulation BCSCs by over expression miR-34a. We found the higher expression change of cytokines profile, such as CXCL11 (I-TAC), CCL11, CXCL10 (IP-10), and the lower expression change of cytokines profile, such as CXCL2 and CXCL12 (SDF-1). The expression of specificity receptors located in NK cell surface, such as CXCR3, CCR5 and CXCR1, which were recognized and bound with the ligands of CXCL11 (I-TAC) and CXCL10 (IP10), and related to the cytotoxic functions of NK cells. These observations suggested that miR-34a could influence NK cells by regulating Notch1 pathway. Enhancement of BCSCs cytotoxic susceptibility to NK cells could finally lead to effective dual suppression to BCSCs and even kill them. Here, NK cells undoubtedly played synergistic and supplemental roles in executing BCSCs.
NK cells exert killing functions through a variety of DNAX-activating protein of 10 kDa (DAP10)-associated receptors. NKG2D was the major DAP10-associated cytotoxicity receptor; NKG2D-NKG2DLs pathway played a vital role in NK cells activation.33 Tian et al proposed the molecular targeted-adoptive cellular immunotherapy, which was based on the biological characteristic of NK cells activation of signaling pathways (NKG2D-NKG2DLs), especially on the basis of the regulation of NKG2DLs.34 NK cells were capable of killing tumor cells or CSCs via receptor-ligand specific recognition and combination. There were both inhibitory receptors and activated receptors on the surface of NK cells. Among them, the value of an activated receptor NKG2D should not be underestimated, some researches indicated that NKG2D and its ligands Rae-1 and/or H60 might play vital roles in graft-versus-tumor (GVT) response.35,36 A number of studies suggested that the human NKG2D could transmit activated signals through DAP10 YxxM base sequence combined with PI3K,37,38 but the regulation mechanism in mice was not entirely clear. In the current study, miR-34a could boost the role of p53 leading to direct change of p53 and PTEN, which might activate the NKG2D-NKG2DLs through combining the PI3K/AKT signal pathway. We found the distinct up-regulation of the activated ligands in both mRNA and protein level by interfering Notch1 or Hes1, however, Notch1 signaling involved in the regulation mechanism needs more research.
In a summary, miR-34a can down-regulate the stemness of BCSCs by targeting Notch1, and promote NK cells' killing function by inducing ligands overexpression of NK cell activated receptors on BCSCs. The emerging dual role of miR-34a in obliterating BCSCs properties might establish a strong rationale for developing miR-34a as a therapeutic agent that targets BCSCs.
Supplementary Material
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Funding
This work was supported by grants from the National Natural Science Foundation of China (NSFC) (Nos.81171975) and Tianjin Institutes for Basic Sciences 15JCYBJC26900.
References
- 1.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 2004; 116:281-97; PMID:14744438; http://dx.doi.org/ 10.1016/S0092-8674(04)00045-5 [DOI] [PubMed] [Google Scholar]
- 2.DeSantis C, Ma J, Bryan L, Jemal A. Breast cancer statistics, 2013. CA 2014; 64:52-62; PMID:24114568; http://dx.doi.org/ 10.1017/S0009840X1300228X [DOI] [PubMed] [Google Scholar]
- 3.Liu C, Kelnar K, Liu B, Chen X, Calhoun-Davis T, Li H, Patrawala L, Yan H, Jeter C, Honorio S, et al.. The microRNA miR-34a inhibits prostate cancer stem cells and metastasis by directly repressing CD44. Nat Med 2011; 17:211-5; PMID:21240262; http://dx.doi.org/ 10.1038/nm.2284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bu P, Chen KY, Chen JH, Wang L, Walters J, Shin YJ, Goerger JP, Sun J, Witherspoon M, Rakhilin N, et al.. A microRNA miR-34a-regulated bimodal switch targets Notch in colon cancer stem cells. Cell Stem Cell 2013; 12:602-15; PMID:23642368; http://dx.doi.org/ 10.1016/j.stem.2013.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ma W, Xiao GG, Mao J, Lu Y, Song B, Wang L, Fan S, Fan P, Hou Z, Li J, et al.. Dysregulation of the miR-34a-SIRT1 axis inhibits breast cancer stemness. Onco Target 2015; 6:10432-44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Corney DC, Hwang CI, Matoso A, Vogt M, Flesken-Nikitin A, Godwin AK, Kamat AA, Sood AK, Ellenson LH, Hermeking H, et al.. Frequent downregulation of miR-34 family in human ovarian cancers. Clin Cancer Res 2010; 16:1119-28; PMID:20145172; http://dx.doi.org/ 10.1158/1078-0432.CCR-09-2642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shi Y, Liu C, Liu X, Tang DG, Wang J. The microRNA miR-34a inhibits non-small cell lung cancer (NSCLC) growth and the CD44hi stem-like NSCLC cells. PloS One 2014; 9:e90022; PMID:24595209; http://dx.doi.org/ 10.1371/journal.pone.0090022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rathod SS, Rani SB, Khan M, Muzumdar D, Shiras A. Tumor suppressive miRNA-34a suppresses cell proliferation and tumor growth of glioma stem cells by targeting Akt and Wnt signaling pathways. FEBS Open Bio 2014; 4:485-95; PMID:24944883; http://dx.doi.org/ 10.1016/j.fob.2014.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang S, Li Y, Gao J, Zhang T, Li S, Luo A, Chen H, Ding F, Wang X, Liu Z. MicroRNA-34 suppresses breast cancer invasion and metastasis by directly targeting Fra-1. Oncogene 2013; 32:4294-303; PMID:23001043; http://dx.doi.org/ 10.1038/onc.2012.432 [DOI] [PubMed] [Google Scholar]
- 10.Kang L, Mao J, Tao Y, Song B, Ma W, Lu Y, Zhao L, Li J, Yang B, Li L. MicroRNA-34a suppresses the breast cancer stem cell-like characteristics by downregulating Notch1 pathway. Cancer Sci 2015; 106:700-8; PMID:25783790; http://dx.doi.org/ 10.1111/cas.12656 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 11.Leong KG, Karsan A. Recent insights into the role of Notch signaling in tumorigenesis. Blood 2006; 107:2223-33; PMID:16291593; http://dx.doi.org/ 10.1182/blood-2005-08-3329 [DOI] [PubMed] [Google Scholar]
- 12.Li Y, Guessous F, Zhang Y, Dipierro C, Kefas B, Johnson E, Marcinkiewicz L, Jiang J, Yang Y, Schmittgen TD, et al.. MicroRNA-34a inhibits glioblastoma growth by targeting multiple oncogenes. Cancer Res 2009; 69:7569-76; PMID:19773441; http://dx.doi.org/ 10.1158/0008-5472.CAN-09-0529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Babiarz JE, Blelloch R. Small RNAs - their biogenesis, regulation and function in embryonic stem cells StemBook Cambridge MA: Harvard Stem Cell Institute, 2008. [PubMed] [Google Scholar]
- 14.Peng GL, Tian Y, Lu C, Guo H, Zhao XW, Guo YW, Wang LQ, Du QL, Liu CP. Effects of Notch-1 down-regulation on malignant behaviors of breast cancer stem cells. J Huazhong University of Science and Technology Medical sciences = Hua zhong ke ji da xue xue bao Yi xue Ying De wen ban = Huazhong keji daxue xuebao Yixue Yingdewen ban 2014; 34:195-200; PMID:24710932; http://dx.doi.org/ 10.1007/s11596-014-1258-4 [DOI] [PubMed] [Google Scholar]
- 15.Diefenbach A, Jamieson AM, Liu SD, Shastri N, Raulet DH. Ligands for the murine NKG2D receptor: expression by tumor cells and activation of NK cells and macrophages. Nat Immunol 2000; 1:119-26; PMID:11248803; http://dx.doi.org/ 10.1038/77793 [DOI] [PubMed] [Google Scholar]
- 16.Wang B, Wang Q, Wang Z, Jiang J, Yu SC, Ping YF, Yang J, Xu SL, Ye XZ, Xu C, et al.. Metastatic consequences of immune escape from NK cell cytotoxicity by human breast cancer stem cells. Cancer Res 2014; 74:5746-57; PMID:25164008; http://dx.doi.org/ 10.1158/0008-5472.CAN-13-2563 [DOI] [PubMed] [Google Scholar]
- 17.Fan X, Chen X, Deng W, Zhong G, Cai Q, Lin T. Up-regulated microRNA-143 in cancer stem cells differentiation promotes prostate cancer cells metastasis by modulating FNDC3B expression. BMC Cancer 2013; 13:61; PMID:23383988; http://dx.doi.org/ 10.1186/1471-2407-13-61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ponti D, Costa A, Zaffaroni N, Pratesi G, Petrangolini G, Coradini D, Pilotti S, Pierotti MA, Daidone MG. Isolation and in vitro propagation of tumorigenic breast cancer cells with stem/progenitor cell properties. Cancer Res 2005; 65:5506-11; PMID:15994920; http://dx.doi.org/ 10.1158/0008-5472.CAN-05-0626 [DOI] [PubMed] [Google Scholar]
- 19.Outtz HH, Wu JK, Wang X, Kitajewski J. Notch1 deficiency results in decreased inflammation during wound healing and regulates vascular endothelial growth factor receptor-1 and inflammatory cytokine expression in macrophages. J Immunol (Baltimore, Md : 1950) 2010; 185:4363-73; http://dx.doi.org/ 10.4049/jimmunol.1000720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sell S. On the stem cell origin of cancer. Am J Pathol 2010; 176:2584-494; PMID:20431026; http://dx.doi.org/ 10.2353/ajpath.2010.091064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Visvader JE, Lindeman GJ. Cancer stem cells in solid tumours: accumulating evidence and unresolved questions. Nat Rev Cancer 2008; 8:755-68; PMID:18784658; http://dx.doi.org/ 10.1038/nrc2499 [DOI] [PubMed] [Google Scholar]
- 22.Yu F, Yao H, Zhu P, Zhang X, Pan Q, Gong C, Huang Y, Hu X, Su F, Lieberman J, et al.. let-7 regulates self renewal and tumorigenicity of breast cancer cells. Cell 2007; 131:1109-23; PMID:18083101; http://dx.doi.org/ 10.1016/j.cell.2007.10.054 [DOI] [PubMed] [Google Scholar]
- 23.Melton C, Judson RL, Blelloch R. Opposing microRNA families regulate self-renewal in mouse embryonic stem cells. Nature 2010; 463:621-6; PMID:20054295; http://dx.doi.org/ 10.1038/nature08725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shimono Y, Zabala M, Cho RW, Lobo N, Dalerba P, Qian D, Diehn M, Liu H, Panula SP, Chiao E, et al.. Downregulation of miRNA-200c links breast cancer stem cells with normal stem cells. Cell 2009; 138:592-603; PMID:19665978; http://dx.doi.org/ 10.1016/j.cell.2009.07.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Esquela-Kerscher A, Slack FJ. Oncomirs - microRNAs with a role in cancer. Nat Rev Cancer 2006; 6:259-69; PMID:16557279; http://dx.doi.org/ 10.1038/nrc1840 [DOI] [PubMed] [Google Scholar]
- 26.Gerhardt DM, Pajcini KV, D'Altri T, Tu L, Jain R, Xu L, Chen MJ, Rentschler S, Shestova O, Wertheim GB, et al.. The Notch1 transcriptional activation domain is required for development and reveals a novel role for Notch1 signaling in fetal hematopoietic stem cells. Genes Dev 2014; 28:576-93; PMID:24637115; http://dx.doi.org/ 10.1101/gad.227496.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tian H, Biehs B, Chiu C, Siebel CW, Wu Y, Costa M, de Sauvage FJ, Klein OD. Opposing activities of Notch and Wnt signaling regulate intestinal stem cells and gut homeostasis. Cell Reports 2015; 11:33-42; PMID:25818302; http://dx.doi.org/ 10.1016/j.celrep.2015.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yamaguchi M, Murakami S, Yoneda T, Nakamura M, Zhang L, Uezumi A, Fukuda S, Kokubo H, Tsujikawa K, Fukada S. Evidence of Notch-Hesr-Nrf2 Axis in Muscle Stem Cells, but Absence of Nrf2 Has No Effect on Their Quiescent and Undifferentiated State. PloS One 2015; 10:e0138517; PMID:26418810; http://dx.doi.org/ 10.1371/journal.pone.0138517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang GC, Xu YH, Chen HX, Wang XJ. Acute lymphoblastic leukemia cells inhibit the differentiation of bone mesenchymal stem cells into osteoblasts in vitro by activating notch signaling. Stem Cells Int 2015; 2015:162410; PMID:26339248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang C, Mo R, Yin B, Zhou L, Liu Y, Fan J. Tumor suppressor microRNA-34a inhibits cell proliferation by targeting Notch1 in renal cell carcinoma. Oncol Letters 2014; 7:1689-94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Park EY, Chang E, Lee EJ, Lee HW, Kang HG, Chun KH, Woo YM, Kong HK, Ko JY, Suzuki H, et al.. Targeting of miR34a-NOTCH1 axis reduced breast cancer stemness and chemoresistance. Cancer Res 2014; 74:7573-82; PMID:25368020; http://dx.doi.org/ 10.1158/0008-5472.CAN-14-1140 [DOI] [PubMed] [Google Scholar]
- 32.Liu C, Kelnar K, Vlassov AV, Brown D, Wang J, Tang DG. Distinct microRNA expression profiles in prostate cancer stem/progenitor cells and tumor-suppressive functions of let-7. Cancer Res 2012; 72:3393-404; PMID:22719071; http://dx.doi.org/ 10.1158/0008-5472.CAN-11-3864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Friese MA, Wischhusen J, Wick W, Weiler M, Eisele G, Steinle A, Weller M. RNA interference targeting transforming growth factor-β enhances NKG2D-mediated antiglioma immune response, inhibits glioma cell migration and invasiveness, and abrogates tumorigenicity in vivo. Cancer Res 2004; 64:7596-603; PMID:15492287; http://dx.doi.org/ 10.1158/0008-5472.CAN-04-1627 [DOI] [PubMed] [Google Scholar]
- 34.Cheng M, Chen Y, Xiao W, Sun R, Tian Z. NK cell-based immunotherapy for malignant diseases. Cell Mol Immunol 2013; 10:230-52; PMID:23604045; http://dx.doi.org/ 10.1038/cmi.2013.10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Champsaur M, Lanier LL. Effect of NKG2D ligand expression on host immune responses. Immunol Rev 2010; 235:267-85; PMID:20536569; http://dx.doi.org/ 10.1111/j.0105-2896.2010.00893.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cerwenka A, Bakker AB, McClanahan T, Wagner J, Wu J, Phillips JH, Lanier LL. Retinoic acid early inducible genes define a ligand family for the activating NKG2D receptor in mice. Immunity 2000; 12:721-7; PMID:10894171; http://dx.doi.org/ 10.1016/S1074-7613(00)80222-8 [DOI] [PubMed] [Google Scholar]
- 37.Tokuyama M, Lorin C, Delebecque F, Jung H, Raulet DH, Coscoy L. Expression of the RAE-1 family of stimulatory NK-cell ligands requires activation of the PI3K pathway during viral infection and transformation. PLoS Pathogens 2011; 7:e1002265; PMID:21966273; http://dx.doi.org/ 10.1371/journal.ppat.1002265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Park YP, Choi SC, Kiesler P, Gil-Krzewska A, Borrego F, Weck J, Krzewski K, Coligan JE. Complex regulation of human NKG2D-DAP10 cell surface expression: opposing roles of the gammac cytokines and TGF-beta1. Blood 2011; 118:3019-27; PMID:21816829; http://dx.doi.org/ 10.1182/blood-2011-04-346825 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.