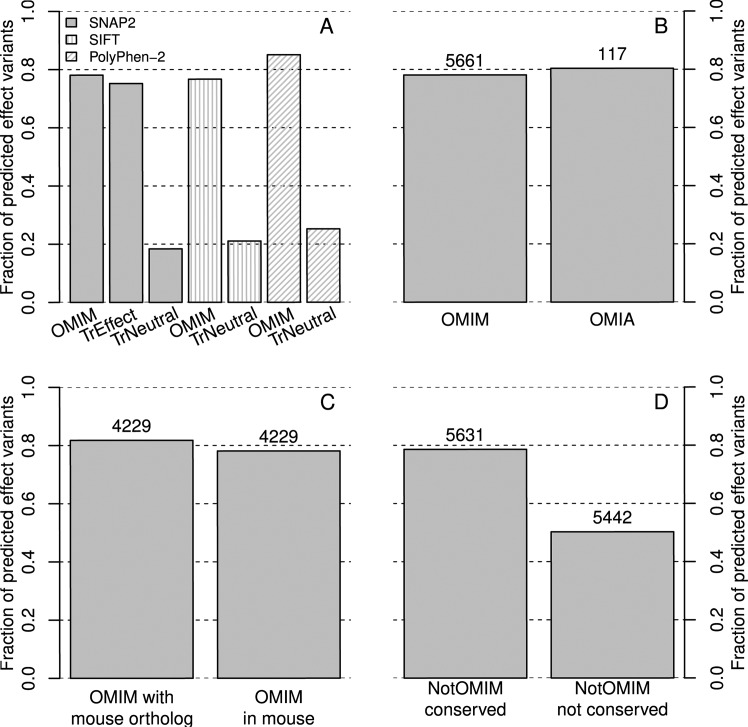
Fig 1. Predictions of SAV effects upon function and disease across species.
The numbers above bars give the number of SAVs in the set. A: Three methods (SNAP2 [16], SIFT [27], PolyPhen-2 [12]) predicted SAV effects upon molecular function (TrEffect/TrNeutral) and upon disease (OMIM). Exclusively for this panel SNAP2 was trained without using disease SAVs from OMIM [5] or HumVar [28]. The SNAP2 version trained exclusively on molecular function clearly captured aspects of OMIM-disease SAVs (leftmost bar OMIM higher than 2nd to the left TrEffect). TrNeutral was the SNAP2 training set of variants without effect. Comparing the bars for TrNeutral and OMIM for each method pointed to differential thresholds: Polyphen-2 correctly predicted more effect in OMIM than SNAP2 but also incorrectly predicted more effect in the neutral data, i.e. simply predicted more effect variants. B: OMIM is repeated from A. SNAP2 captured disease signals in humans and animals at similar levels. OMIA contained disease SAVs from animals other than mouse and rat (mostly dog and cattle). C: SNAP2 predicted OMIM SAVs with less effect in mouse orthologs than in human. Left bar (OMIM with mouse ortholog): SNAP2 predictions for the subset of all 4,229 OMIM SAVs for which we found a mouse ortholog. Right bar (OMIM in mouse): SNAP2 predictions when putting the human SAV into the mouse sequence. D: Disease variants happen in non-random positions. Left bar (NotOMIM conserved): in each protein with an OMIM SAV, we predicted the effect of all SAVs with a level of sequence conservation ≥ that of the OMIM variant. Right bar (NotOMIM not conserved): predictions for SAVs in non-OMIM positions with conservation < that of the OMIM SAV. Obviously, OMIM SAVs were very well conserved.