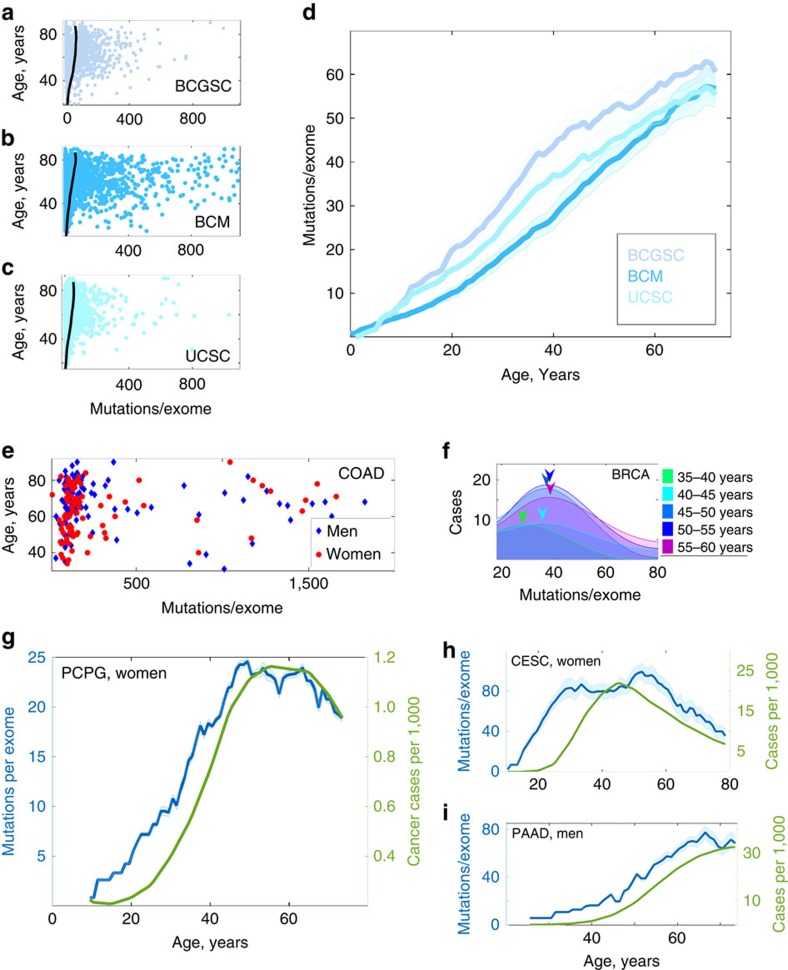
Figure 1. Accumulation of mutations with age and cancer incidence.
(a) Somatic mutation distribution and combined mutation load for gender-nonspecific cancers (ESCA, LIHC, PAAD, PCPG, SARC, STAD and UVM in both men and women) sequenced by Canada's Michael Smith Genome Sciences Centre (BCGSC). Each point represents a particular cancer sample. Black line denotes behaviour of the median of the distribution with age. (b) The same for samples sequenced by Baylor College of Medicine (BCM); cancers include ESCA, LIHC, LGG, PAAD, PCPG, STAD, THCA and UVM. (c) The same for samples sequenced by the University of California-Santa Cruz (UCSC); cancers include ESCA, KIRP, LIHC, PAAD, PCPG, SARC and UVM. (d) Behaviour of the mutation load distribution medians with age for distributions depicted in a–c. (e) Mutation load distribution for patients with colon adenocarcinoma (COAD), samples sequenced by BCM, IlluminaGA pipeline. Cancers in men are shown in blue, and in women in red. (f) Dynamics of the mutation load distribution P(N, t) of somatic mutation counts N with age t for breast adenocarcinoma (BRCA) in women. Samples sequenced by Washington University, IlluminaGA human-curated pipeline. The actual histograms of mutation loads are approximated by 8 degree polynomials. For each age cohort, the distribution peak is denoted by a coloured arrow. (g) Somatic mutation load versus cancer incidence (cases per 1,000 capita) for pheochromocytoma and paraganglioma (PCPG) cancers in women; samples sequenced by Broad Institute, IlluminaGA automated variant-calling pipeline. Cancer incidences (green) and age-related somatic mutation accumulation pattern (blue). (h) The same for cervical squamous cell carcinoma and endocervical adenocarcinoma (CESC). Samples sequenced by BCGSC, IlluminaHiSeq automated pipeline. (i) The same for pancreatic adenocarcinoma (PAAD) in men. Samples sequenced by BCM, IlluminaGA automated pipeline. Results for other cancers, datacenters and variant-calling pipelines are presented in Supplementary Figs 1–16. Errors are s.d., calculated using bootstrapping as described in Methods.