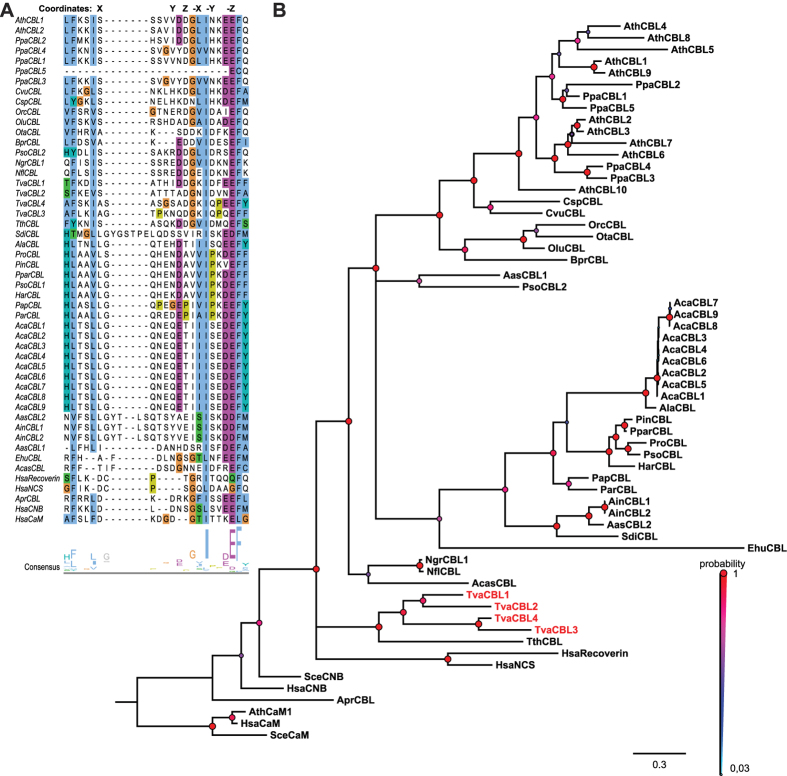
Figure 1. Newly identified Ca2+ sensors are phylogenetically closely related to CBLs from Arabidopsis thaliana and share characteristic sequence features with CBL-type proteins.
(A) Multiple Sequence Alignment (MSA) of the first EF-hand of Arabidopsis CBL1 and CBL2 with Ca2+ sensors from other species. MSA is visualized by Jalview Version 2 and colored by clustal X color scheme according to conservation (B) Phylogenetic relationship of CBL and CBL-related Ca2+ sensors. Depicted is a 50%-majority-rule consensus tree based on Bayesian statistics (MrBayes). CBLs and CBL-like candidates cluster on one branch next to their nearest relatives in metazoan (NCS, Recoverin). Values above branches indicate Bayesian posteriori probabilities determined by MrBayes phylogenetic tree inference. The tree was rooted with the CaM branch. Accession numbers are provided in Supplementary Table S1; Ath: Arabidopsis thaliana, Apr: Auxenochlorella protothecoides, Ain: Aphanomyces invadans, Acas: Acanthamoeba castellanii, Aas: Aphanomyces astaci, Ala: Albugo laibachii, Aca: Albugo candida, Bpr: Bathycoccus prasino, Csp: Chlorella sp., Cvu: Chlorella vulgaris, Ehu: Emiliania huxleyi, Har: Hyaloperonospora arabidopsidis, Ngr: Naegleria gruberi, Nfl: Naegleria fowleri, Ota: Ostreococcus tauri, Olu: Ostreococcus lucimarinus, Orc: Ostreococcus RCC 809, Ppa: Physcomitrella patens, Pin: Phytophthora infestans, Pra: Phytophthora ramorum, Pso: Phytophthora sojae, Pap: Pythium aphanidermatum, Par: Pythium arrhenomanes, Ppar: Phytophthora parasitica, Tth: Tetrahymena thermophila, Tva: Trichomonas vaginalis.