Abstract
Background
Bloodstream infections (BSI) are life-threatening emergencies. Identification of the common pathogens and their susceptibility patterns is necessary for timely empirical intervention.
Methods
We conducted a 4-year retrospective analysis of blood cultures from all patients excluding neonates at the Korle-Bu Teaching hospital, Ghana, from January 2010 through December 2013. Laboratory report data were used to determine BSI, blood culture contamination, pathogen profile, and antimicrobial resistance patterns.
Results
Overall, 3633 (23.16 %) out of 15,683 blood cultures were positive for various organisms. Pathogen-positive cultures accounted for 1451 (9.3 %, 95 % CI 8.5–9.8 %). Infants recorded the highest true blood culture positivity (20.9 %, n = 226/1083), followed by the elderly (13.3 %, n = 80/601), children (8.9 %, n = 708/8000) and adults (7.2 %, n = 437/6000) (p = 0.001 for Marascuilo’s post hoc). Overall occurrence of BSI declined with increasing age-group (p = 0.001) but the type of isolates did not vary with age except for Citrobacter, Escherichia coli, Klebsiella, Salmonella, and Enterococcus species. Gram negative bacteria predominated in our study (59.8 %, n = 867/1451), but the commonest bacterial isolate was Staphylococcus aureus (21.9 %, n = 318/1451)—and this trend run through the various age-groups. From 2010 to 2013, we observed a significant trend of yearly increase in the frequency of BSI caused by cephalosporin-resistant enterobacteria (Chi square for trend, p = 0.001). Meropenem maintained high susceptibility among all Gram-negative organisms ranging from 96 to 100 %. Among Staphylococcus aureus, susceptibility to cloxacillin was 76.6 %.
Conclusion
Our study shows a significantly high blood culture positivity in infants as compared to children, adults and the elderly. There was a preponderance of S. aureus and Gram-negative bacteria across all age-groups. Meropenem was the most active antibiotic for Gram-negative bacteria. Cloxacillin remains a very useful anti-staphylococcal agent.
Electronic supplementary material
The online version of this article (doi:10.1186/s12941-016-0163-z) contains supplementary material, which is available to authorized users.
Keywords: Ghana, Bloodstream, Infections, Infants, Adults, Antibiotic susceptibility
Background
Developing countries, especially in Africa, have a disproportionate burden of global bacterial bloodstream infections (BSI). This contributes significantly to morbidity, mortality, and increased health care costs [1–3]. The potential outcomes of bloodstream infections are such that treatment is often empirical, due to delays in performing and receiving blood culture results. In developing countries, this is further worsened by the unavailability of well-resourced microbiology facilities [4]. Empirical antibiotics used in such instances are usually based on international guidelines and not guided by local susceptibility data [5]. The rise in antibiotic resistance worldwide means that the likelihood of inadequate therapy in BSI is heightened, with increased chances of poor therapeutic outcomes [6, 7]. Knowledge of the local antibiotic resistance profile of bacteria increases the probability of selecting effective empirical therapy [2, 8]. In a recent study on the cost-effectiveness of bloodstream infection surveillance for the management of sepsis in low-resource settings, Penno et al. [5] showed that antibiotic prescribing, based on local antibiogram is able to save 534 additional lives per 100,000 patients. The finding suggests that it is crucial to monitor emerging trends in resistance at the local level to support clinical management. However, data on antimicrobial resistance, especially regarding trends in BSI, remain scarce in Ghanaian healthcare settings including Korle-Bu Teaching Hospital (KBTH).
Published data describing BSI in KBTH are limited to short study periods [9] and have mainly focused on children [10]. Using a large collection of blood culture data, we have examined trends in pediatric and adult BSI over 4 years at KBTH; determined blood culture yields and contamination rate; and analyzed the antibiotic susceptibility profiles of the associated organisms, with emphasis on multidrug resistant phenotypes of public health concern.
Methods
Study design
This was a hospital based retrospective analysis of blood culture reports from January 2010 to December 2013 at the Korle-Bu Teaching Hospital in Ghana, West Africa. We followed guidelines for the reporting of studies conducted using observational routinely-collected health data (RECORD) [11]. The KBTH is a 2000-bed tertiary teaching hospital with about 200 admissions per day [12]. The central outpatient department records about 29,757 patients per month [12]. The hospital covers all medical specialties and provides referral healthcare services to an estimated population of 24 million Ghanaians.
Study settings and population
We collated blood culture results of patients older than 28 days, from the bacteriology unit of the Microbiology Laboratory in KBTH. Data on BSI of patients ≤28 days old will be published elsewhere. The bacteriology unit of KBTH processes over 40,000 clinical cultures annually. To obtain a good representation of the repertoire of BSI isolates occurring at a tertiary health facility, we have included all blood culture results of patients primarily receiving healthcare at the KBTH (n = 15,072) or secondarily referred to the laboratory from other health facilities for microbiological investigations (n = 611). We included data for the period January 2010 to December 2013. Data prior to this period were not properly archived and therefore difficult to access.
Selection, review and extraction of data from laboratory reports
A data entry team comprising three bacteriologists and a physician who assisted with the medical reconciliation of data conducted a manual work-through of laboratory paper records to select blood cultures conducted between 2010 and 2013. Selected blood culture reports were reviewed and extracted into a computerized database (Statistical Package for Social Sciences, Version 20.0) as the primary data source. We had no independent means of validating laboratory results, thus reports were excluded from entry on the basis of incomplete data if results regarding the positivity or otherwise of blood cultures were inconclusive. The quality of abstracted data was assessed by an independent reviewer in a standardized manner by comparing paper records to the electronic entries. Disagreements between data were resolved by consensus between the reviewer and the data entry team. Extracted information included patient demographics, bacterial and fungal isolates, BSI categorization, and antibiogram of isolates. Reports for patients aged ≤28 days were not considered for analysis. True positive cultures were those with identified bacteria plus corresponding susceptibility results, or yeast. Organisms, including Micrococcus species, Bacillus species, and diphtheroids were classified as contaminants. For the majority of patients, only a single blood culture was submitted per infection episode. Where duplicate cultures were submitted, and the same organism was isolated within 14 days of the previous culture, then the latter isolate was excluded except when there was variation in antibiotic susceptibility pattern.
Data source/methods of laboratory assessment for blood cultures
For patients with suspected sepsis, local guidelines recommended the inoculation of 1–3 mL (for paediatric patients; however, for older children, larger blood inoculums of 10 mL were encouraged) and 8–10 mL (for adults) directly into Bactec® culture vials (Becton–Dickinson, USA). Routinely at the laboratory, cultures were processed with the BACTEC 9240 blood culture system (Becton–Dickinson, NJ, USA) according to manufacturer’s instructions. Where bacterial growth was detected in vials, Gram-stains were performed; and subcultures were typically made onto appropriate media based on Gram-stain results. Bacterial isolates were identified using routine biochemical methods. Bacteria speciation was done with the BBL® Crystal identification system (Becton–Dickinson, NJ, USA). For positive fungal blood cultures, organisms were identified on the basis of morphology. As part of regular practice at the laboratory, consultant microbiologists evaluated all positive blood cultures to categorize isolates as contaminants or true pathogens. Susceptibility testing for bacterial pathogens were conducted using the Kirby-Bauer disc diffusion method with antibiotic discs; and these tests were interpreted according to guidelines by the Clinical and Laboratory Standards Institute (CLSI) [13].
Antibiotic resistance phenotypes
We assessed the occurrence of six epidemiologically important bacterial pathogens: vancomycin resistant Enterococcus species (VRE) [based on in vitro susceptibility to vancomycin disk (30 μg)], methicillin resistant Staphylococcus aureus (MRSA) [based on in vitro susceptibility to cefoxitin disk (30 μg)], penicillin resistant streptococci (PRS) [based on in vitro susceptibility to ampicillin disk (10 μg)], cephalosporin resistant enterobacteria (Ceph-R Ent) [based on in vitro susceptibility to cefotaxime disk (30 μg)], multi-drug resistant Pseudomonas species (MDR Ps.) and multi-drug resistant Acinetobacter species (MDR Act). A multidrug resistant (MDR) phenotype was defined, relative to the panel of antibiotics reported for each bacteria, as in vitro non-susceptibility to ≥1 agent in ≥3 antimicrobial classes: penicillins, cephalosporins, β-lactamase inhibitor combinations, carbapenems, tetracyclines, folate pathway inhibitors, glycopeptides, fluoroquinolones, chloramphenicol, aminoglycosides, and macrolides [14].
Data analysis
Data analysis was performed with the Statistical Package for Social Sciences, Version 20.0. The distribution of BSI and resistance profiles of bacterial isolates were assessed using descriptive methods. Blood culture positivity and contamination levels were calculated by dividing positive blood cultures and contaminants respectively by the total number of blood cultures submitted. Susceptibility tests reports were classified as resistant or susceptible, and the data expressed as the proportion of susceptible isolates out of the total valid test results for the specific antibiotic and pathogen. Missing data were excluded from analysis. Study data were compared across four patient age categories: infants (>28 days–1 year), children (>1–15 years), adults (>15–65 years), and the elderly (>65 years). The Chi square analysis with Marascuilo’s post hoc tests for multiple comparisons was used to compare categorical data within patient groups. A Chi square test for linear trend was used to assess changes in categorical variables over the study period. Continuous data were compared using Students’ t test with analyses of variance (ANOVA) for multiple comparisons.
Ethical considerations
During the data extraction process, we de-identified blood culture reports to ensure complete obscurity from laboratory archives. Arbitrary numbers were assigned to all reported isolates. Patients’ clinical data were not reviewed for analysis. Owing to the retrospective study design, and given the anonymized nature of our data, patients’ consent for use of the laboratory records could not be obtained. Ethical approval for isolates and laboratory data was not required as the study was regarded as part of routine surveillance measures for infection control. Study findings are freely available to the hospital for the formulation of guidelines for treatment of BSI.
Results
Figure 1 shows the flow diagram for data collation and outcome. Overall, a total of 15,683 blood cultures were performed for infants (n = 1082), children (n = 8003), adults (n = 5987), and the elderly (n = 601). Males comprised 53 % (n = 8312) of patients. The median age of the study population was 31.6 years (range 29 days–69 years). About 3633 (23.16 %) blood cultures were positive: 1451 (9.3 %, 95 % CI 8.5–9.8) were considered true pathogens and 2182 (13.9 %) considered contaminants. The rate of BSI was 92.5 per 1000 blood cultures submitted to the laboratory, with no significant variation across the years under review (2010, 82.4 per 1000 blood cultures; 2011, 94.2 per 1000 blood cultures; 2012, 87.2 per 1000 blood cultures; 2013, 90.3 per 1000 blood cultures: p = 0.0841 for Marascuilo’s post hoc). The median age of patients with BSI was 13.5 years (range 29–81 years). Chi square for linear trend analysis showed that the overall occurrence of BSI declined with increasing age-group (p = 0.001), but most of the isolates did not vary with age (Table 1). The highest true blood culture positivity was observed among infants (20.9 %, n = 226/1083), followed by the elderly (13.3 %, n = 80/601), children (8.9 %, n = 708/8000) and adults (7.2 %, n = 437/6000) (p = 0.001 for Marascuilo’s post hoc). The proportion of blood cultures positive for various pathogens remained similar (p = 0.412 for Chi square test) between patients receiving care at KBTH (9.3 %, n = 1407/15,072) and those referred to the laboratory from other health facilities (7.6 %, n = 44/611). In the proceeding analysis, we have disregarded comparing results between the two sources of blood cultures because there were few true positive cultures from the latter.
Fig. 1.
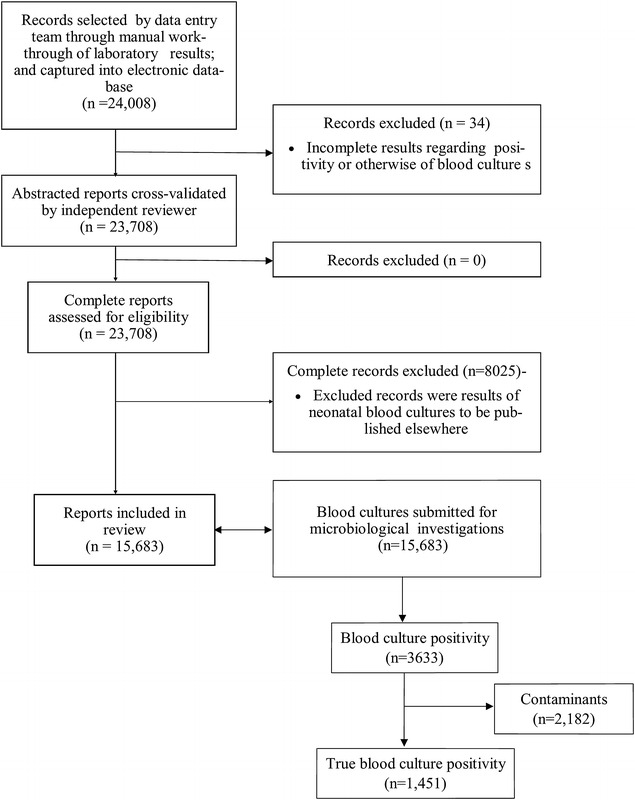
Flow diagram for data collation and results outcome. Overall, 24,042 laboratory records were reviewed, 15,683 of which belonged to patients >28 days and were eligible for inclusion. Of the 15,683 blood cultures submitted, 1451 were positive with various pathogens
Table 1.
Distribution of BSI isolates across age groups
| Organism (n, %) | Infants (A) (29 days–1 year) | Children (B) (1–15 years) | Adults (C) (15–65 years) | Elderly (D) (>65 years) | Post hoc comparison |
 trend trend |
|---|---|---|---|---|---|---|
| GNB (n = 867, 59.8 %) | 123 (54.4) | 407 (57.5) | 265 (65.4) | 52 (65.0) | A = B = C = D | 0.002 |
| Enterobacteria (n = 640, 44.1 %) | 81 (35.8) | 312 (44.1) | 203 (46.4) | 44 (55.0) | A < D = B = C | 0.001 |
| Citrobacter sp. (n = 148, 10.2 %) | 14 (6.2) | 58 (8.1) | 65 (13.0) | 11 (13.7) | A = B = C = D | 0.001 |
| E. coli (n = 118, 8.1 %) | 8 (3.5) | 27 (3.8) | 60 (13.7) | 23 (28.7) | A = B < C = D* | 0.001 |
| Enterobacter sp. (n = 105, 7.2 %) | 14 (6.2) | 53 (7.5) | 32 (7.3) | 6 (7.5) | A = B = C = D | 0.731 |
| Klebsiella sp. (n = 63, 4.3 %) | 16 (7.1) | 30 (4.2) | 14 (3.2) | 3 (3.8) | A = B = C = D | 0.037 |
| Proteus sp. (n = 9, 0.6 %) | 2 (0.8) | 3 (0.4) | 4 (0.9) | 0 | A = B = C = D | 0.668 |
| Providencia sp. (n = 4, 0.4 %) | 1 (0.4) | 1 (0.1) | 2 (0.5) | 0 | A = B = C = D | 0.738 |
| P. agglomerans (n = 2, 0.1 %) | 0 | 1 (0.1) | 1 (0.2) | 0 | A = B = C = D | 0.992 |
| Salmonella sp. (169, 11.6 %) | 23 (9.7) | 127 (17.9) | 18 (4.3) | 1 (1.3) | B > C = A > D* | 0.001 |
| Serratia sp. (n = 1, 0.1 %) | 0 | 0 | 1 (0.2) | 0 | A = B = C = D | 0.755 |
| Other coliform (n = −21, 1.4 %) | 3 (1.3) | 12 (1.7) | 6 (1.4) | 0 | A = C = D < B | 0.429 |
| Non-fermentative bacilli (n = 223, 15.3 %) | 42 (18.5) | 92 (12.9) | 81 (18.5) | 8 (10.0) | A = B = C = D | 0.818 |
| Acinetobacter sp. (n = 120, 8.3 %) | 24 (10.6) | 56 (7.9) | 36 (8.2) | 4 (5.0) | A = B = C = D | 0.173 |
| Pseudomonas sp. (n = 101, 7.0 %) | 18 (7.9) | 35 (4.9) | 44 (10.0) | 4 (5.0) | A = B < C = D* | 0.404 |
| S. paucimobilis (n = 1, 0.1 %) | 0 | 1 (0.1) | 0 | 0 | A = B = C = D | 0.334 |
| S. maltophilia (n = 1, 0.1 %) | 0 | 0 | 1 (0.2) | 0 | A = B = C = D | 0.759 |
| Other gram negatives (n = 4, 0.2 %) | 0 | 3 (0.4) | 1 (0.2) | 0 | A = B = C = D | 0.738 |
| Chromobacter sp. (n = 1, 0.1 %) | 0 | 0 | 1 (0.2) | 0 | A = B = C = D | 0.759 |
| N. meningitides (n = 2, 0.2 %) | 0 | 3 (0.4) | 0 | 0 | A = B = C = D | 0.350 |
| GPB (n = 563, 36.5 %) | 102 (45.1) | 289 (40.7) | 146 (25.9) | 26 (32.5) | C < A = B = D | 0.0001 |
| Enterococcus sp. (n = 43, 0.3 %) | 8 | 24 (3.4) | 10 (22.8) | 1 (1.3) | A = B = C = D | 0.139 |
| S. aureus (n = 318, 21.9 %) | 60 (26.5) | 148 (20.9) | 99 (22.6) | 11 (13.8) | A = B = C = D | 0.092 |
| S. epidermidis (n = 67, 4.6 %) | 11 (4.8) | 42 (5.9) | 12 (2.7) | 2 (2.6) | A = B = C = D | 0.047 |
| S. agalactiae (n = 1, 0.1 %) | 1 (0.4) | 0 | 0 | 1 (1.3) | A = B = C = D | 0.984 |
| S. pneumoniae (n = 35, 2.4 %) | 6 (2.6) | 23 (3.2) | 3 (0.6) | 3 (3.8) | A = B < C = D* | 0.159 |
| S. pyogenes (n = 3, 0.2 %) | 0 | 1 (0.1) | 1 (0.2) | 1 (1.3) | A = B = C = D | 0.217 |
| S. viridans (n = 48, 3.3 %) | 10 (4.4) | 23 (3.2) | 9 (8.9) | 6 (7.5) | A = B = C = D | 0.739 |
| Other Streptococcus sp. (n = 48, 3.3 %) | 7 (3.1) | 28 (4.0) | 12 (2.7) | 1 (1.3) | A = B = C = D | 0.243 |
| Fungi (n = 21, 1.3 %) | 2 (0.8) | 12 (1.7) | 5 (1.1) | 2 (2.6) | A = B = C = D | 0.596 |
| Aspergillus sp. (n = 3, 0.2 %) | 0 | 2 (0.2) | 0 | 1 (1.3) | A = B = C = D | 0.588 |
| Candida sp. (n = 17, 1.2 %) | 2 (0.8) | 10 (1.4) | 4 (0.9) | 1 (1.3) | A = B = C = D | 0.816 |
| Cryptococcus sp. (n = 1, 0.1 %) | 0 (0) | 0 | 1 (0.2) | 0 | A = B = C = D | 0.759 |
| True blood culture positivity (n = 1451, 9.3 %) | 226 (20.9 %) | 708 (8.9 %) | 437 (7.3 %) | 80 (13.3 %) | A > D > B > C* | <0.001 |
Sp., species; GNB, gram negative bacteria; GPB, gram positive bacteria; E. coli, Escherichia coli; P. agglomerans, Pantoea agglomerans; S. paucimobilis, Sphingomonas paucimobilis; S. maltophilia, Stenotrophomonas maltophilia; N. meningitis, Neisseria meningitis; S. aureus, Staphylococcus aureus; S. epidermidis, Staphylococcus epidermidis; S. agalactiae, Streptococcus agalactiae; S. pneumoniae, Streptococcus pneumoniae; S. pyogenes, Streptococcus pyogenes; S. viridans, Streptococcus viridans;  trend, Chi square for linear trend; Post hoc, comparison compares the proportion of bloodstream infections among age-groups
trend, Chi square for linear trend; Post hoc, comparison compares the proportion of bloodstream infections among age-groups
* Comparisons with significant (p < 0.05) differences
Distribution of isolates
Gram-negative bacteria accounted for 59.8 % (n = 867/1451) of BSI pathogens, whilst Gram-positives and fungi accounted for 38.8 % (n = 563/1451) and 1.4 % (n = 21/1451) respectively (p = 0.001 for Marascuilo’s post hoc)—and the trend was similar within each age-group. Staphylococcus aureus was the most isolated organism (21.9 %, n = 318/1451). The majority of Gram-negative bacteria belonged to the family Enterobacteriaceae (73.9 %, n = 640/867), with Salmonella (11.1 %) and Citrobacter (10.2 %) species being the second and third most isolated pathogens (Table 1).
Antibiotic susceptibility
Our results show high levels of ampicillin resistance among Gram-negative (Table 2) and Gram-positive (Table 3) organisms. Susceptibility among the enterobacteria ranged from 0 % among Proteus and Klebsiella species to 36.1 % in Salmonella species. There was 100 % susceptibility among Neisseria meningitides isolates. Among Gram-positive organisms, susceptibility ranged from 7.7 % in Enterococcus species to 100 % in Streptococcus pyogenes. Among the aminoglycosides, Gram-negative organisms maintained high susceptibility to amikacin than to gentamicin. For Pseudomonas species, susceptibility of 86.9 and 67.2 % were respectively recorded for amikacin and gentamicin, whilst for Citrobacter species, susceptibility of 84.7 and 33.1 % were noted. In this study, moderate to high level susceptibility of Gram-negative organisms to quinolones was recorded, exemplified by 64.9 and 69.6 % susceptibility of Citrobacter species, as well as 100 and 97.6 % susceptibility of Salmonella species, to ciprofloxacin and levofloxacin respectively. Most enterobacteria showed low to moderate (20–43 %) susceptibility to cefotaxime. High percentage susceptibility were however recorded for Salmonella (95.5 %). Meropenem retained high susceptibility among all Gram-negative organisms, ranging from 96 % among Pseudomonas species to 100 % among Citrobacter species. Among Staphylococcus aureus, susceptibility to cloxacillin over the period was 76.6 %, cefuroxime 48.5 %, erythromycin 70 %, co-trimoxazole 24 %, gentamicin 68 % and vancomycin 100 %. Isolates of Streptococcus pneumoniae were moderately susceptible to penicillin (32.5 %) and ampicillin (46.7), but highly susceptible to erythromycin (84.2 %) and vancomycin (100 %).
Table 2.
Percentage susceptibility of gram negative pathogens
| Organism | Amp | Am/cl | Gen | Amk | Cip | Lev | Cef | Ctx | Mem | Cot | Tet | Chl |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Citrobacter sp. | 6/120 (5) | 8/33 (24.2) | 44/133 (33.1) | 100/118 (84.7) | 46/64 (71.9) | 19/27 (70.4) | 34/136 (25) | 46/142 (32.4) | 148/148 | 19/113 (16.8) | 15/103 (14.6) | 11/96 (11.5) |
| Serratia sp. | 0/1 | 1/1 | 1/1 | 1/1 | 1/1 | 1/1 | 1/1 | 0/1 | 1/1 | 1/1 | ||
| E. coli | 2/86 (2.3) | 5/40 (12.5) | 40/86 (46.5) | 67/87 (77) | 25/66 (37.9) | 9/31 (29) | 31/104 (29.8) | 49/112 (43.8) | 118/118 (100) | 6/79 (7.6) | 6/69 (8.7) | 8/71 (11.3) |
| Enterobacter sp. | 2/91 (2.2) | 2/29 (6.9) | 27/86 (31.4) | 68/89 (76.4) | 37/57 (64.9) | 16/23 (69.6) | 19/98 (19.4) | 22/100 (22) | 104/105 (99) | 14/81 (17.3) | 7/71 (9.9) | 7/64 (10.9) |
| Klebsiella sp. | 0/56 (0) | 3/18 (16.7) | 11/52 (21.2) | 43/53 (81.1) | 20/33 (60.1) | 12/19 (63.2) | 9/59 (15.3) | 12/60 (20) | 62/63 (98.4) | 7/53 (13.2) | 3/45 (6.7) | 6/44 (13.6) |
| Proteus sp. | 0/6 (0) | 2/4 (50) | 5/7 (71.4) | 5/6 (83.3) | 4/5 (80) | 1/2 (50) | 5/9 (5.6) | 6/9 (66.7) | 9/9 | 1/6 (16.7) | 0/3 (0) | 1/5 (20) |
| Providencia sp. | 0/3 | 3/4 (75) | 4/4 | 1/1 | 1/4 (25) | 2/4 (50) | 4/4 | 0/4 | 1/3 (33.3) | 0/5 | ||
| P. agglomerans | 0/2 | 1/2 (50) | 2/2 | 0/2 | 0/2 | 2/2 | 0/2 | 0/2 | ||||
| Salmonella sp. | 48/133 (36.1) | 35/46 (76.1) | 125/133 (94) | 131/133 (98.5) | 96/96 (100) | 40/41 (97.6) | 130/156 (83.3) | 154/162 (95.1) | 169/169 | 59/122 (48.3) | 35/106 (33) | 29/111 (26.1) |
| Serratia sp. | 0/1(0) | 1/1 | 1/1 | 1/1 | 1/1 | 1/1 | 1/1 | 0/1 | 1/1 | 1/1 | ||
| Acinetobacter sp. | 4/56 (7.1) | 6/16 (37.5) | 25/55 (45.5) | 43/57 (75.4) | 18/25 (72) | 10/17 (58.8) | 17/66 (25.8) | 16/70 (22.9) | 116/120 (96.7) | 12/51 (23.5) | 3/50 (6) | 6/50 (12) |
| Pseudomonas sp. | 45/67 (67.2) | 53/61 (86.9) | 41/51 (80.4) | 39/49 (79.6) | 97/101 (96) | |||||||
| S. maltophilia | 0/1 | 0/1 | 1/1 | 1/1 | ||||||||
| S. paucimobilis | 1/1 | 1/1 | 1/1 | 1/1 | 1/1 | 1/1 | 0/1 | 0/1 | ||||
| Chromobacter sp. | 0/1 | 1/1 | 1/1 | 0/1 | 1/1 | 1/1 | ||||||
| N. meningitidis | 1/1 | 1/1 | 2/3 (66.7) | 2/3 (66.7) | 3/3 | 0/1 | 0/1 |
Amp ampicillin, Am/cl amoxicillin clavulanic acid, Gen gentamicin, amk amikacin, cip ciprofloxacin, Lev levofloxacin, Cef cefuroxime, ceft cefotaxime, Mem meropenem, Cot cotrimoxazole, Tet tetracycline, Chl chloramphenicol, sp species, E. coli Escherichia coli, P. agglomerans Pantoea agglomerans, S. paucimobilis Sphingomonas paucimobilis,S. maltophilia Stenotrophomonas maltophilia,N. meningitis Neisseria meningitis
Table 3.
Percentage susceptibility of gram positive pathogens
| Organisms | Pen | Amp | Amc/cl | Gen | Cef | Ox | Vanc | Ery | Tet | Ctx | Cot | Chl |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Staphylococcus epidermidis | 3/6 (50) | 4/4 (100) | ||||||||||
| Staphylococcus aureus | 20/293 (6.8) | 26/260 (10) | 42/60 (70) | 170/250 (68) | 141/291 (48.5) | 226/295 (76.6) | 144/144 (100) | 111/182 (70) | 59/242 (24.4) | 89/233 (38.2) | ||
| Enterococcus sp. | 1/38 (2.6) | 3/39 (7.7) | 15/17 (88.2) | 14/33 (42.4) | ||||||||
| Streptococcus pyogenes | 3/3 (100) | 2/2 (100) | 2/2 (100) | 3/3 (100) | 2/2 (100) | 2/2 (100) | 1/1 (100) | |||||
| Streptococcus viridans | 4/45 (8.9) | 7/42 (16.6) | 5/13 (34.5) | 16/40 (.4) | 19/20 (95) | 5/20 (25) | 28/40 (70) | 4/25 (16) | ||||
| Streptococcus agalactiae | 0/1 (0) | 0/1 (0) | 0/1 (0) | 0/1 (0) | 0/1 (0) | |||||||
| Streptococcus pneumoniae | 10/31 (32.5) | 14/30 (46.7) | 10/10 (100) | 27/39 (69.2) | 12/12 (100) | 16/19 (84.2) | 32/32 (100) | 4/19 (21.1) | ||||
| Streptococcus sp. | 3/41 (7.3) | 6/42 (14.3) | 6/9 (66.7) | 20/43 (46.5) | 24/27 (88.9) | 15/28 (53.8) | 12/23 (52.2) | 12/35 (34.3) |
Data not available for blank cells
Pen penicillin, Amp ampicillin, Am/cl amoxicillin clavulanic acid, Gen gentamicin, Cef cefuroxime, Cot cotrimoxazole, Tet tetracycline, Chl chloramphenicol, Ctx cefotaxime, Chl chloramphenicol
Trends in antibiotic susceptibility patterns
In Fig. 2, we assessed antibiotic susceptibility patterns over 4 years, looking out for significant trends. There was a significant rise in susceptibility to penicillin (p = 0.001), ampicillin (p = 0.000), cefuroxime (0.0000), cloxacillin (p = 0.0000), cotrimoxazole (p = 0.012), cefotaxime (p = 0.0197) and chloramphenicol (p = 0.001). Figure 3 shows the occurrence of some epidemiologically important antibiotic resistance phenotypes: VRE, MRSA, PRS, Cef-R Ent, MDR Ps, MDR Act, and all MDRs. From 2010 to 2013, we observed a significant trend towards increasing frequency of BSI caused by Cef-R Ent (Chi square for trend, p = 0.001). No other resistant phenotype varied with years.
Fig. 2.
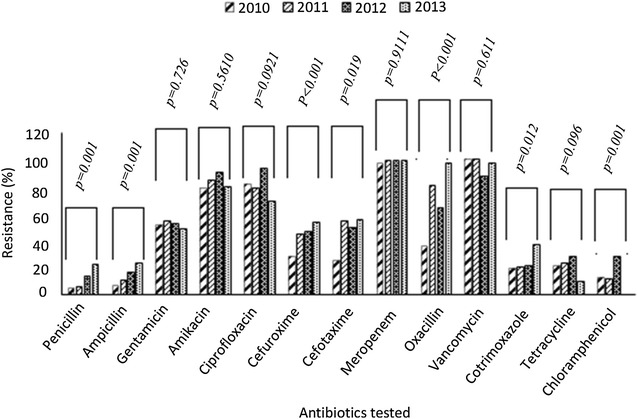
Trends in antibiotic susceptibility patterns over 4 years (2010 through to 2013). p values calculated for Mantel–Haenszel Chi square for linear trends
Fig. 3.
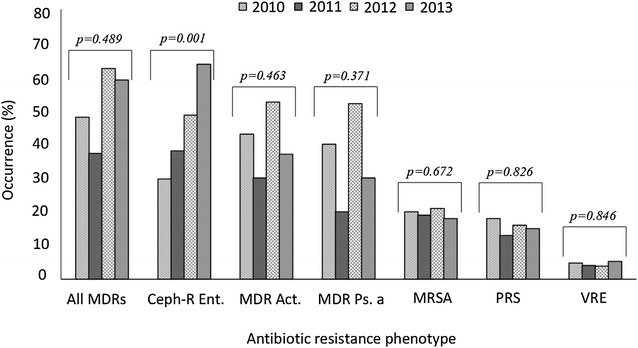
Antimicrobial resistance phenotypes over 4 years (2010 through to 2013). p values calculated for Mantel–Haenszel Chi square for linear trends. MDRs multidrug resistant bacteria, VRE vancomycin resistant Enterococci, MRSA methicillin resistant Staphylococcus aureus, PRS penicillin resistant Streptococci, Cef-R Ent cephalosporin resistant Enterobacteriaceae, MDR Ps multidrug resistant Pseudomonas aeruginosa, MDR Act multidrug resistant Acinetobacter species
Discussion
Bloodstream infections accounted for 9.3 % of blood cultures and varied significantly within age-groups, where the highest prevalence was recorded among patients at the extremes of ages: the elderly (13.3 %) and infants (20.9 %). The reasons for this distribution is unclear, as risk factors for developing BSI were not assessed in this study. Nevertheless, infants were significantly more likely to have BSI compared with patients aged >3 years. There may be several reasons for this association. Infants are likely to have frequent exposure to healthcare environments or participate in activities that predispose them to microbial contamination. They also have poor skin integrity and an immature immune system [15–19].
In keeping with previous studies from Ghana [20, 21], Gram-negative bacteria predominated in our study, but the majority of bacterial findings were S. aureus, which also predominated in other studies from Guinea-Bissau [22], Nigeria [16] and Malawi [23]. We believe that the aetiology of BSI is changing, so that BSI are due largely to Gram-negative bacteria, but with a significant contribution from Gram-positive isolates, mostly Staphylococcus species. Other studies in Africa have documented this phenomenon, attributing the predominance of Gram-positive bacteria (mostly S. pneumoniae and S. aureus) to community-acquired infections [24–26] and Gram-negative bacteria (predominantly E. coli and K. pneumoniae) to healthcare related infections [27]. We are unable to determine which BSI were healthcare related or community-acquired because of insufficient data on inpatient and outpatient status. The presence of Streptococcus pneumoniae as a cause of BSI was uncommon in our study. This could partially be attributable to pneumococcal vaccine introduced in January 2012. Immunization of infants with the vaccine has led not only to major declines in childhood invasive pneumococcal sepsis but also to a reduction in infections among adults due to herd immunity [27]. It is the experience in KBTH that Salmonella species constitute a prominent pathogen in BSI, with a preponderance of non-typhoidal salmonellae over typhoidal salmonellae. Children under 5 years bear the brunt of the disease [28]. These observations are generally consistent with findings from other African studies, particularly in malaria-endemic regions, where Salmonella species have predominated in BSI [29, 30].
Favoured by of their low toxicity, broad spectrum and comparatively high effectiveness, the cephalosporin antibiotics are widely used in Ghana. A worrying observation is the increasing isolation of cephalosporin resistant Enterobacteriaceae over the 4-year review period. The enterobacteria constituted over 40 % of the bloodstream pathogens. Given that the variety of class C cephalosporinases (AmpCs) and extended spectrum β-lactamases (ESBLs) remain the principal mechanisms for cephalosporin resistance worldwide [31–33], the high levels of cephalosporin resistance observed in our study may be compatible with results from other studies in Ghana, demonstrating high levels of ESBLs (>45 %) among the enterobacteria [31, 34].
Based on the low frequency of fungi and the dominance of both GPB and GNB, it may be necessary for clinicians to consider suitable empiric regimens that provide adequate cover for both bacterial groups. Cefotaxime, gentamicin, ciprofloxacin and amoxicillin plus clavulanic acid, in various combinations, have been the predominant empiric therapies for bloodstream infections in Ghana [35]. We have demonstrated here and in previous studies that the general antibiogram of the GNB and GPB reveal an overall high resistance to these routinely used drugs [31, 32]. Our observations are in consonance with other studies in Ghana and at Korle-Bu teaching Hospital, which found high resistance to ampicillin, gentamicin and cefotaxime among bacteria isolates of various infections [31, 32, 36]. Carbapenem was the most active antibiotic against Gram-negative infections. Meanwhile, cloxacillin remained a very useful anti-staphylococcal agent due the relatively low prevalence of MRSA.
This study has several limitations. Being retrospective, we were unable to ascertain the standardization of techniques for blood cultures and laboratory procedures for individual patients. The study is also unable to evaluate the accuracy of bacteria isolates classified as contaminants in laboratory records. As the study lacks comprehensive clinical data or information on recent antibiotic use and/or hospitalization, our data on the occurrence of BSI should be interpreted with caution. Finally, our study was performed in a single center, and cannot be generalized to the entire patient population in Ghana. Our study nonetheless provides relevant data which may be used to guide empirical therapy, change antibiotic prescription policy in our institution or serve as a baseline for future research. We recommend future studies to ascertain the changing eligibility of our findings over time beyond the review years reported in this study.
Conclusion
Our study shows that blood culture positivity was significantly higher in infants compared to children, adults and the elderly; with a preponderance of S. aureus and Gram-negative bacteria in all age-groups. The findings presented here suggest the need to review empiric antibiotic options in these patient populations, given the relatively low susceptibility of the bacterial isolates to many routinely used antibiotics.
Authors’ contributions
AKL conceived the study; participated in its design, coordination, and collation of laboratory data; and helped to draft the manuscript. NON participated in the study design, coordination and collation of laboratory data; performed the statistical analysis; and helped to draft the manuscript. JEML, GAM participated in collation of laboratory data and review of the manuscript. NOA participated in the drafting and review of the manuscript. All authors read and approved the final manuscript.
Acknowledgements
This research/paper has been supported by the University of Ghana—Carnegie Next Generation of Academics in Africa Project with funding from the Carnegie Corporation of New York. We are grateful to the staff of the Microbiology Department of the Korle-Bu Teaching Hospital for their support. We thank Dr. Benjamin Jella Visser and (Department of Tropical Medicine, Infectious Diseases at Academic Medical Center, Tergooi, Netherlands) and Professor George Obeng-Adjei (Center for tropical clinical pharmacology and therapeutics, Ghana) for reviewing the manuscript.
Competing interests
The authors declare that they have no competing interests.
Availability of data and materials
The authors do not have the permission of the health institution to share the raw data in publicly available repositories. The RECORD checklist of items, extended from the STROBE statement that should be reported in this study using routinely collected health data for observational studies is attached as Additional file 1.
Funding
The study received no specific grant.
Abbreviations
- µg
microgram
- ANOVA
analysis of variance
- BSI
blood stream infections
- CI
confidence interval
- CLSI
Clinical and Laboratory Standards Institute
- GNB
gram-negative bacteria
- GPB
gram-positive bacteria
- KBTH
Korle-Bu Teaching Hospital
- MDR
multidrug resistance
- SPSS
statistical package for social sciences
- NJ
New Jersey
- USA
United States of America
- VRE
vancomycin resistant Enterococcus species
- MRSA
methicillin resistant Staphylococcus aureus
- PRS
penicillin resistant streptococci
- Ceph-R Ent
cephalosporin resistant enterobacteria
- MDR
multidrug resistant
- MDR Ps
multi-drug resistant Pseudomonas species
- MDR Act
multi-drug resistant Acinetobacter species
Additional file
10.1186/s12941-016-0163-z The RECORD statement—checklist of items, extended from the STROBE statement that should be reported in observational studies using routinely collected health data.
Footnotes
Noah Obeng-Nkrumah and Appiah-Korang Labi contributed equally to this work
Contributor Information
Noah Obeng-Nkrumah, Email: successfulnoahforchrist@yahoo.com.
Appiah-Korang Labi, Email: appiahl@yahoo.com.
Naa Okaikor Addison, Email: nokaikor@gmail.com.
Juliana Ewuramma Mbiriba Labi, Email: juliemills05@yahoo.com.
Georgina Awuah-Mensah, Email: gawuah@chs.edu.gh.
References
- 1.Okeke IN, Klugman KP, Bhutta ZA, Duse AG, Jenkins P, O’Brien TF, Pablos-Mendez A, Laxminarayan R. Antimicrobial resistance in developing countries. Part II: strategies for containment. Lancet Infect Dis. 2005;5:568–580. doi: 10.1016/S1473-3099(05)70217-6. [DOI] [PubMed] [Google Scholar]
- 2.Reynolds R, Potz N, Colman M, Williams A, Livermore D. Antimicrobial susceptibility of the pathogens of bacteraemia in the UK and Ireland 2001–2002: the BSAC bacteraemia resistance surveillance programme. J Antimicrob Chemother. 2004;53:1018–1032. doi: 10.1093/jac/dkh232. [DOI] [PubMed] [Google Scholar]
- 3.Diekema DJ, Beekmann SE, Chapin KC, Morel KA, Munson E, Doern GV. Epidemiology and outcome of nosocomial and community-onset bloodstream infection. J Clin Microbiol. 2003;41:3655–3660. doi: 10.1128/JCM.41.8.3655-3660.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Petti CA, Polage CR, Quinn TC, Ronald AR, Sande MA. Laboratory medicine in Africa: a barrier to effective health care. Clin Infect Dis. 2006;42:377–382. doi: 10.1086/499363. [DOI] [PubMed] [Google Scholar]
- 5.Penno EC, Baird SJ, Crump JA. Cost-effectiveness of surveillance for bloodstream infections for sepsis management in low-resource settings. Am J Trop Med Hyg. 2015;93:850–860. doi: 10.4269/ajtmh.15-0083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blomberg B, Manji KP, Urassa WK, Tamim BS, Mwakagile DS, Jureen R, Msangi V, Tellevik MG, Holberg-Petersen M, Harthug S, Maselle SY, Langeland N. Antimicrobial resistance predicts death in Tanzanian children with bloodstream infections: a prospective cohort study. BMC Infect Dis. 2007;7:43. doi: 10.1186/1471-2334-7-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cosgrove SE. The relationship between antimicrobial resistance and patient outcomes: mortality, length of hospital stay, and health care costs. Clin Infect Dis. 2006;42(Supplement 2):S82–S89. doi: 10.1086/499406. [DOI] [PubMed] [Google Scholar]
- 8.Kamga HLF, Njunda AL, Nde PE, Assob JCN, Nsagha DS, Weledji P. Prevalence of septicaemia and antibiotic sensitivity pattern of bacterial isolates at the University Teaching Hospital, Yaoundé, Cameroon. Afr J Clin Exp Microbiol. 2011;12(1):2–8. [Google Scholar]
- 9.Enweronu-Laryea CC, Newman MJ. Changing pattern of bacterial isolates and antimicrobial susceptibility in neonatal infections in Korle Bu Teaching Hospital, Ghana. East Afr Med J. 2007;84:136–140. doi: 10.4314/eamj.v84i3.9516. [DOI] [PubMed] [Google Scholar]
- 10.Wilkens J, Newman MJ, Commey JO, Seifert H. Salmonella bloodstream infection in Ghanaian children. Clin Microbiol Infect. 1997;3:616–620. doi: 10.1111/j.1469-0691.1997.tb00467.x. [DOI] [PubMed] [Google Scholar]
- 11.Benchimol EI, Smeeth L, Guttmann A, Harron K, Moher D, Petersen I, Sørensen HT, von Elm E, Langan SM. The reporting of studies conducted using observational routinely-collected health data (RECORD) statement. PLoS Med. 2015;12:1–22. doi: 10.1371/journal.pmed.1001885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Annual Report Korle-Bu Teaching Hospital. 2010.
- 13.Wayne PA. Clinical and Laboratory Standard Institute (CLSI): performance standards for antimicrobial susceptibility testing. Twenty Second Inf Suppl. 2012;32(3):1–278. [Google Scholar]
- 14.Magiorakos A, Srinivasan A, Carey RB, Carmeli Y, Falagas ME, Giske CG, Harbarth S, Hindler JF. bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect. 2012;18:268–281. doi: 10.1111/j.1469-0691.2011.03570.x. [DOI] [PubMed] [Google Scholar]
- 15.Ugas MA, Cho H, Trilling GM, Tahir Z, Raja HF, Ramadan S, Jerjes W, Giannoudis PV. Central and peripheral venous lines-associated blood stream infections in the critically ill surgical patients. Ann Surg Innov Res. 2012;6:8. doi: 10.1186/1750-1164-6-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hojsak I, Strizić H, Mišak Z, Rimac I, Bukovina G, Prlić H, Kolaček S. Central venous catheter related sepsis in children on parenteral nutrition: a 21-year single-center experience. Clin Nutr. 2012;31:672–675. doi: 10.1016/j.clnu.2012.02.006. [DOI] [PubMed] [Google Scholar]
- 17.Pronovost PJ, Goeschel CA, Colantuoni E, Watson S, Lubomski LH, Berenholtz SM, Thompson DA, Sinopoli DJ, Cosgrove S, Sexton JB, Marsteller JA, Hyzy RC, Welsh R, Posa P, Schumacher K, Needham D. Sustaining reductions in catheter related bloodstream infections in Michigan intensive care units: observational study. BMJ. 2010;340:c309. doi: 10.1136/bmj.c309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Downes KJ, Metlay JP, Bell LM, McGowan KL, Elliott MR, Shah SS. Polymicrobial bloodstream infections among children and adolescents with central venous catheters evaluated in ambulatory care. Clin Infect Dis. 2008;46:387–394. doi: 10.1086/525265. [DOI] [PubMed] [Google Scholar]
- 19.Costello JM, Clapper TC, Wypij D. Minimizing complications associated with percutaneous central venous catheter placement in children: recent advances. Pediatr Crit Care Med. 2013;14:273–283. doi: 10.1097/PCC.0b013e318272009b. [DOI] [PubMed] [Google Scholar]
- 20.Acquah S, Quaye L, Sagoe K, Ziem J, Bromberger P, Amponsem AA. Susceptibility of bacterial etiological agents to commonly-used antimicrobial agents in children with sepsis at the Tamale Teaching Hospital. BMC Infect Dis. 2013;13:89. doi: 10.1186/1471-2334-13-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Groß U, Amuzu SK, de Ciman R, Kassimova I, Groß L, Rabsch W, Rosenberg U, Schulze M, Stich A, Zimmermann O. Bacteremia and antimicrobial drug resistance over time, Ghana. Emerg Infect Dis. 2011;17:1879–1882. doi: 10.3201/edi1710.110327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Isendahl J, Manjuba C, Rodrigues A, Xu W, Henriques-normark B, Giske CG, Nauclér P. Prevalence of community-acquired bacteraemia in Guinea-Bissau : an observational study. BMC Infect Dis. 2014;14:1–9. doi: 10.1186/s12879-014-0715-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Makoka MH, Miller WC, Hoffman IF, Cholera R, Gilligan PH, Kamwendo D, Malunga G, Joaki G, Martinson F, Hosseinipour MC. Bacterial infections in Lilongwe, Malawi: aetiology and antibiotic resistance. BMC Infect Dis. 2012;12:67. doi: 10.1186/1471-2334-12-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mckay R, Bamford C, Chb MB, Microbiology M, Town C, Africa S. Community- versus healthcare-acquired bloodstream infections at Groote Schuur Hospital, Cape Town, South Africa. S Afr Med J. 2015;105:363–369. doi: 10.7196/SAMJ.8183. [DOI] [PubMed] [Google Scholar]
- 25.Prasad N, Murdoch DR, Reyburn H, Crump JA. Etiology of severe febrile illness in low- and middle-income countries : a systematic review. PLos One. 2015;10:1–25. doi: 10.1371/journal.pone.0127962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hill PC, Onyeama CO, Ikumapayi UNA, Secka O, Ameyaw S, Simmonds N, Donkor SA, Howie SR, Tapgun M, Corrah T, Adegbola RA. Bacteraemia in patients admitted to an urban hospital in West Africa. BMC Infect Dis. 2007;20:1–8. doi: 10.1186/1471-2334-7-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Laupland KB, Sciences CH, Hospital RI. Incidence of bloodstream infection: a review of population-based studies. Clin Microbiol Infect. 2013;19:492–500. doi: 10.1111/1469-0691.12144. [DOI] [PubMed] [Google Scholar]
- 28.Labi AK, Obeng-Nkrumah N, Addison NO, Donkor ES. Salmonella blood stream infections in a tertiary care setting in Ghana. BMC Infect Dis. 2014;14:3857. doi: 10.1186/s12879-014-0697-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.MacLennan CA. Out of Africa: links between invasive nontyphoidal Salmonella disease, typhoid fever, and malaria. Clin Infect Dis. 2013;58(5):648–650. doi: 10.1093/cid/cit803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Phoba M-F, De Boeck H, Ifeka BB, Dawili J, Lunguya O, Vanhoof R, Muyembe J-J, Van Geet C, Bertrand S, Jacobs J. Epidemic increase in Salmonella bloodstream infection in children, Bwamanda, the Democratic Republic of Congo. Eur J Clin Microbiol Infect Dis. 2014;33:79–87. doi: 10.1007/s10096-013-1931-8. [DOI] [PubMed] [Google Scholar]
- 31.Obeng-Nkrumah N, Twum-Danso K, Krogfelt KA, Newman MJ. High levels of extended-spectrum beta-lactamases in a major teaching hospital in Ghana: the need for regular monitoring and evaluation of antibiotic resistance. Am J Trop Med Hyg. 2013;89:960–964. doi: 10.4269/ajtmh.12-0642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Newman MJ, Frimpong E, Donkor ES, Opintan JA, Asamoah-Adu A. Resistance to antimicrobial drugs in Ghana. Infect Drug Resist. 2011;4:215–220. doi: 10.2147/IDR.S21769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bush K. Bench-to-bedside review: the role of β-lactamases in antibiotic-resistant gram-negative infections. Crit Care. 2010;14:224. doi: 10.1186/cc8892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hackman HK, Brown CA, Twum-Danso K. Antibiotic resistance profile of non-extended spectrum beta-lactamase-producing Escherichia coli and Klebsiella pneumoniae in Accra, Ghana. J Biol Agric Healthc. 2014;4:12–16. [Google Scholar]
- 35.Ministry of Health. Republic of Ghana Standard Treatment Guidelines, 6th edn. Accra, Ghana: Ghana National Drugs programme; 2010.
- 36.Ayisi LA, Adu-Sarkodie Y. Extended-spectrum-beta-lactamase (ESBL) production among Escherichia coli and Klebsiella species in Kumasi, Ghana. J Nat Sci Res. 2015;5:81–86. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The authors do not have the permission of the health institution to share the raw data in publicly available repositories. The RECORD checklist of items, extended from the STROBE statement that should be reported in this study using routinely collected health data for observational studies is attached as Additional file 1.


